
Jinxin Phaedo Chen
@chenphaedo
PhD student @BicroLabs at @karolinskainst and @scilifelab; previous BIOPICian @PKU1898; Developing next-gen sequencing techniques; 3D genome organization
ID: 1344531460200415234
http://www.jphaedochen.com/ 31-12-2020 06:31:28
2,2K Tweet
499 Takipçi
856 Takip Edilen

Inspired by Cajal, the Hansel lab's new work in eLife - the journal provides a comparative histological analysis of human and mouse Purkinje cell morphology. Following our 2023 study, we detail differences of scale and kind between the neurons of the two species 🧵👇 elifesciences.org/articles/105013




Thank you Weill Cornell Medicine newsroom for featuring our Molecular Cell study with Dr. Fine on the identification and perturbation of three-Dimensional Gene Hubs in Brain Cancer. news.weill.cornell.edu/news/2025/04/t…




"Native nucleosomes know where to go!” Our Condense-seq paper is now published in Nature! 📄 nature.com/articles/s4158… I’m deeply grateful to my PhD advisor Taekjip Ha and our incredible collaborators The Muir Lab , Bin Zhang , Erika Pearce, and Ben Garcia lab!!





Can blocking “jumping genes” treat diseases and aging? Learn more: scim.ag/4iPW2p7 News from Science



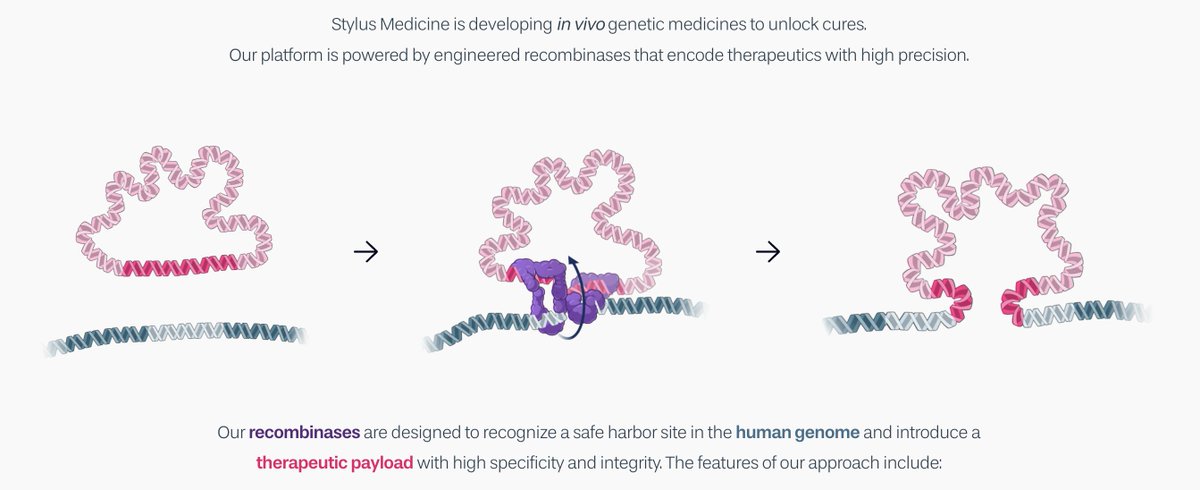
Stylus originated from a wonderful scientific collaboration discovering serine DNA recombinases with Ami Bhatt, Lacra Bintu, and Michael C Bassik spearheaded by star researchers Matt Durrant Alison Fanton Josh Tycko A huge thanks also to Cosmas Giallourakis and Emily Minkow for their


Thank you Itai Yanai and the everyone for such exceptional workshops! It's valuable in expanding my perspectives and mindsets. These workshops are essential for researchers. I'll more enjoy the epiphanies and mystic encounters in my contradictory data now 🐘☕!




