
Ariana Huebner
@amihuebner
ID: 917361125695676417
09-10-2017 12:08:30
27 Tweet
138 Takipçi
262 Takip Edilen

Wearing our #UnityBand on #WorldCancerDay in support of all the patients taking part in the studies that will allow us to #BeatCancerSooner CRUKLondon Science and Innovation at Cancer Research UK UCL Cancer Institute


Our last paper exploring the importance of whole-genome doubling during cancer evolution. Nicholas McGranahan Charles Swanton

Our paper, a collaboration with Enkhtsetseg Munkhbaatar, Philipp Jost and Nicholas McGranahan is published in Nature Communications! We highlight the frequent occurrence of MCL-1 gains in lung adenocarcinoma and explore the potential of MCL-1 as a therapeutic target. nature.com/articles/s4146…

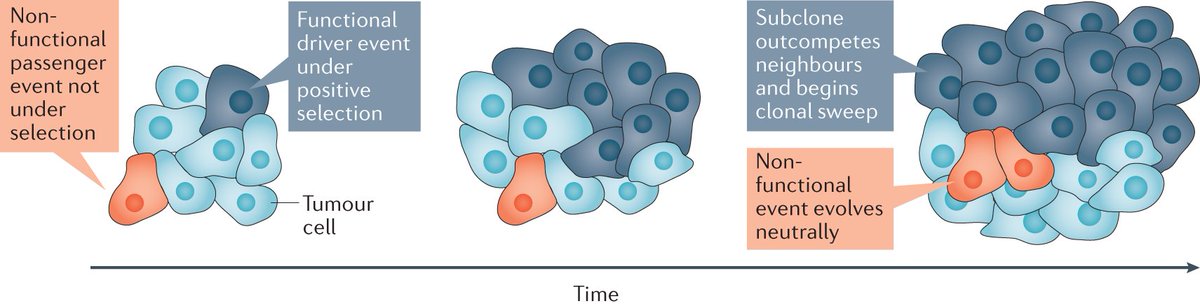
You always had trouble understanding tumour evolution? Our new SnapShot in Cell highlights the key concepts of tumour evolution and drivers of diversity for you. Thanks for the great collaboration Ariana Huebner and Nicholas McGranahan! cell.com/cell/fulltext/…

Excited to share our Cell SnapShot on Tumor Evolution which sheds light on key concepts of tumor evolution and drivers of diversity. Plus it looks great as a poster! Michelle Dietzen Nicholas McGranahan cell.com/cell/fulltext/…

Enjoyed Saturday's #AACR21 sessions on #cancerevolution? Check out this recent Review by JRM Black James Black & N McGranahan Nicholas McGranahan: Genetic and non-genetic clonal diversity in cancer evolution, go.nature.com/3qUIHB4


Extremely proud of Ariana Huebner - passing her viva with flying colours (never in doubt!)...and many thanks to her examiners Florian Markowetz @NischalanP UCL Cancer Institute CRUKCOLcentre CRUK Lung Centre #cancerevolution #cancermetastasis




5 years in the making - happy to see this work from my PhD published! Thanks to James Black and @Rodrigo23374678 for the collaboration and Nicholas McGranahan for his supervision. Thread below 👇

📢The RNA single base substitution signatures we described in #TRACERx are online at COSMIC v3.4! Check them out if you're studying RNA editing in cancer Led by @CarlosEcolEvol with Nicholas McGranahan Charles Swanton Link: cancer.sanger.ac.uk/signatures/rna… Paper: nature.com/articles/s4158…


Our automated method for reconstructing tumor phylogenies, CONIPHER, is finally published! Many thanks to Kristiana Grigoriadis and all co-authors. This tool has been invaluable for processing the recent TRACERx primary and metastasis tumors and we hope it will be for others as well!

Patient-derived xenograft (PDX) models allow cancer researchers to study tumour tissue from patients in mice. Our CRUK Lung Centre paper Nature Communications describes 48 new PDX models and highlights the importance of intratumor heterogeneity in the development of cancer models (1/9)


My Swanton Lab postdoc paper is now out Nature Communications. It was a huge privilege to work with the TRACERx team. Thanks to Ariana Huebner and Dave Pearce, as well as everyone else UCL Cancer Institute, The Francis Crick Institute and CRUK Manchester Institute who made this happen.

Today our study of the association between replication timing alterations and mutation acquisition during cancer evolution has been published in Nature Communications nature.com/articles/s4146…

My main PhD project is finally out 🎉 it was such an honour to investigate replication timing with Haoran Zhai 🥳 many thanks for all the great support to Nicholas McGranahan Nnennaya Kanu and Charles Swanton and all the great scientists that helped us along the way 🫶🏽

Together with Charles Swanton, delighted to share that our new tool, MHC Hammer - which can detect different types of HLA disruption - mutation, loss, RNA repression and alt. splicing - is now out in #NatureGenetics and available to use…nature.com/articles/s4158…... 🧵below

🚀 Our paper introducing ImmuneLENS, a new tool that measures T and B cell fractions from WGS data. This builds on our previous method, T cell ExTRECT, that used a signal from V(D)J recombination to measure T cells. Out today in Nature Genetics doi.org/10.1038/s41588… 🧵👇
