
Ivan Sovic
@ivansovic
Genomics enthusiast with substantial experience in seq alignment, de novo assembly and consensus algorithms, developing professional production-class software.
ID: 433591486
10-12-2011 19:18:10
1,1K Tweet
632 Takipçi
375 Takip Edilen







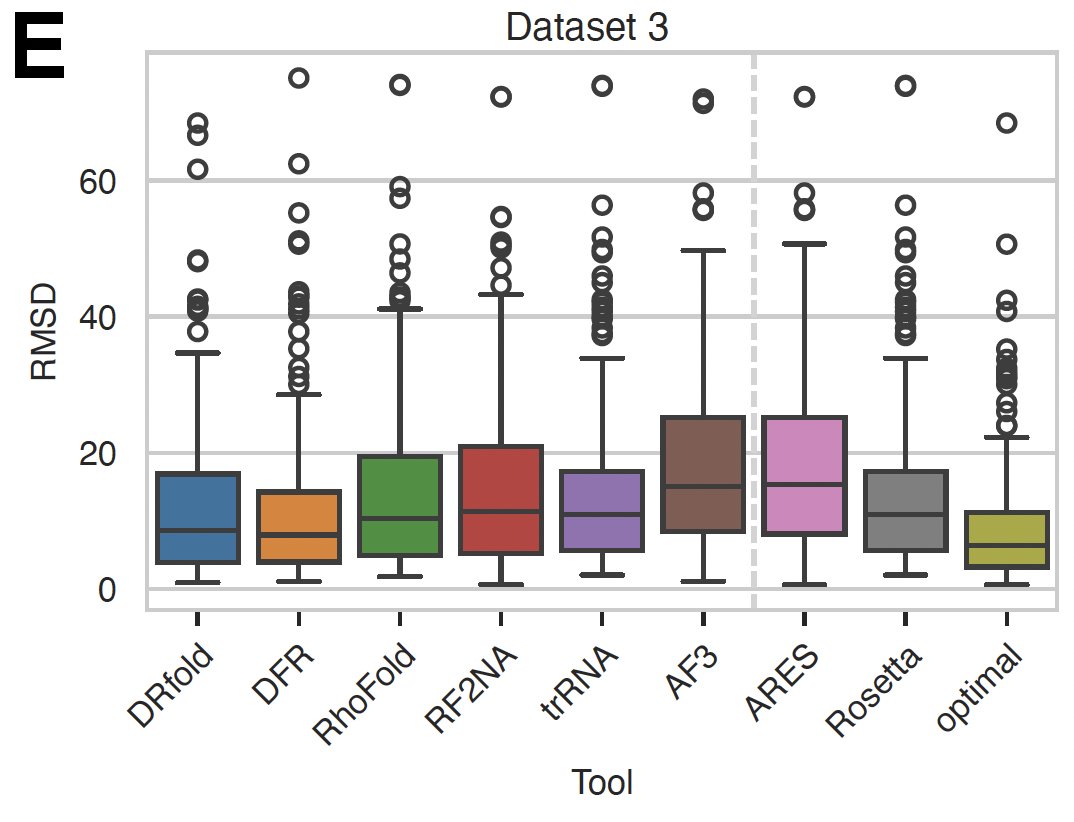
Ivona Martinović Bryan Hooi Selection of the best-predicted structures using Rosetta and ARES. The correlation between trRosettaRNA (trRNA) and Rosetta is not surprising, but look at Alfafold3 and ARES !! #ai #RNA #structure #prediction


A comparative approach that predicts any modifications in RNA (or DNA) using raw Oxford Nanopore signals. Although simple, It outperforms the current SOTA. academic.oup.com/nar/advance-ar… w/ Ivan Vujaklija Siniša Biđin, Marin Volarić, Sara Bakić, Zhe Li, Jianjun Liu and Roger Foo

Our hitchhiking paper is out at genomebiology.biomedcentral.com/articles/10.11… - a great collaborative effort with Prasadms Josipa Lipovac, FilipTomas13 JJ Liu. Briefly, Oxford Nanopore is enough for high-quality human genomes. Recently, HERRO showed that even UL is sufficient. 1/3






wow, turns out even investors want Laravel for JS. We had to sprinkle a bit of AI story on top to make them buy it, but it worked 🤞 Joke aside, Martin Sosic and I are beyond excited to share this news. We still can't believe we get to build the next-generation full-stack web

Join our team - Mile Sikic, Lovro Vrček, Ivona Martinović, Tin Vlašić - at the AI for nucleic acids workshop happening next Monday 28 April at Singapore Expo. Check out: ai4na-workshop.github.io

GNNome was published in Genome Research! This is a novel paradigm for de novo genome assembly based on GNNs. Without explicitly implementing any simplification strategies, it can achieve results comparable or higher than other SOTA tools. Paper, code, and overview are 👇 [1/8]
![Lovro Vrček (@lovrovrcek) on Twitter photo GNNome was published in <a href="/genomeresearch/">Genome Research</a>! This is a novel paradigm for de novo genome assembly based on GNNs. Without explicitly implementing any simplification strategies, it can achieve results comparable or higher than other SOTA tools. Paper, code, and overview are 👇 [1/8] GNNome was published in <a href="/genomeresearch/">Genome Research</a>! This is a novel paradigm for de novo genome assembly based on GNNs. Without explicitly implementing any simplification strategies, it can achieve results comparable or higher than other SOTA tools. Paper, code, and overview are 👇 [1/8]](https://pbs.twimg.com/media/GqAirreawAMb9Oy.jpg)

Our publication in Genome Research presents a new paradigm in genome de novo assembly - using AI instead of classical algorithms. Here, we replace the Layout phase with Graph Neural Networks

I am happy to share our new preprint introducing MADRe - a pipeline for Metagenomic Assembly-Driven Database Reduction, enabling accurate and computationally efficient strain-level metagenomic classification. Mile Sikic, Riccardo Vicedomini, Krešimir Križanović 🔗biorxiv.org/content/10.110… 1/9


