
Ioannis Sarropoulos
@ioansarr
EMBO postdoc @teichlab @SCICambridge and @sangerinstitute / single-cell genomics and gene regulation / PhD in evodevo genomics @kaessmannlab @UniHeidelberg
ID: 963762263206264834
14-02-2018 13:10:04
218 Tweet
381 Followers
758 Following

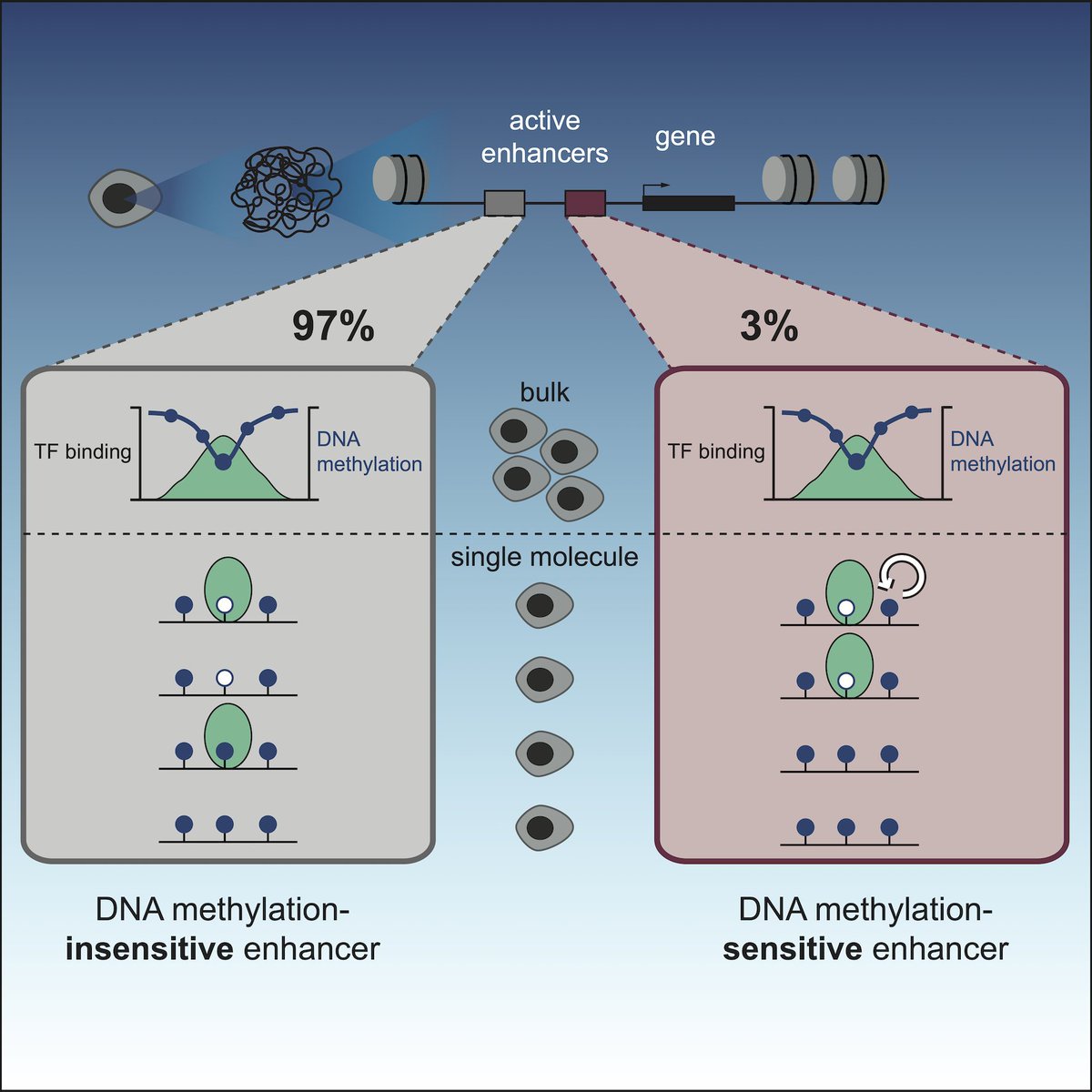
Happy to announce that our story on #DNAmethylation at #enhancers is out today in Molecular Cell! tinyurl.com/smc5yccy It was a fantastic PhD journey in the lab of @arnaud_kr @EMBL. Follow the 🧵 to find out whether #DNAmethylation is important for enhancer activity: (1/9)



We're thrilled to announce that Rasa Elmentaite Rasa Elmentaite, a former TeichLab PhD student, phenomenal young scientist, has been awarded the prestigious Beddington Medal by @TheBSDB for her groundbreaking work in human gut development. 🧬🏅 Read more: bsdb.org/2023/09/04/202…

Our study on the lamprey neural cell type atlas and the origins of the vertebrate brain, led by @frnclmn and Francisca Hervas-Sotomayor, is now out in NatureEcoEvo! nature.com/articles/s4155…


Our study on "Sex-biased gene expression across mammalian organ development and evolution” is out now in Science Magazine! science.org/doi/10.1126/sc…

Excited to share our work in Science Magazine on the evolution and development of sex differences. This study was led by Leticia Rodríguez-Montes and co-supervised by Henrik Kaessmann Kaessmann Lab and I. The Francis Crick Institute 🤝 ZMBH - Center for Molecular Biology Heidelberg Uni Heidelberg Check the 🧵 for more 👇 science.org/doi/10.1126/sc…

Very happy to have my first paper out 💃! Thanks lot to Margarida Cardoso Moreira and Kaessmann Lab for their great support and guidance, and to all coauthors. It’s been an awesome journey to work with you on this project😊.


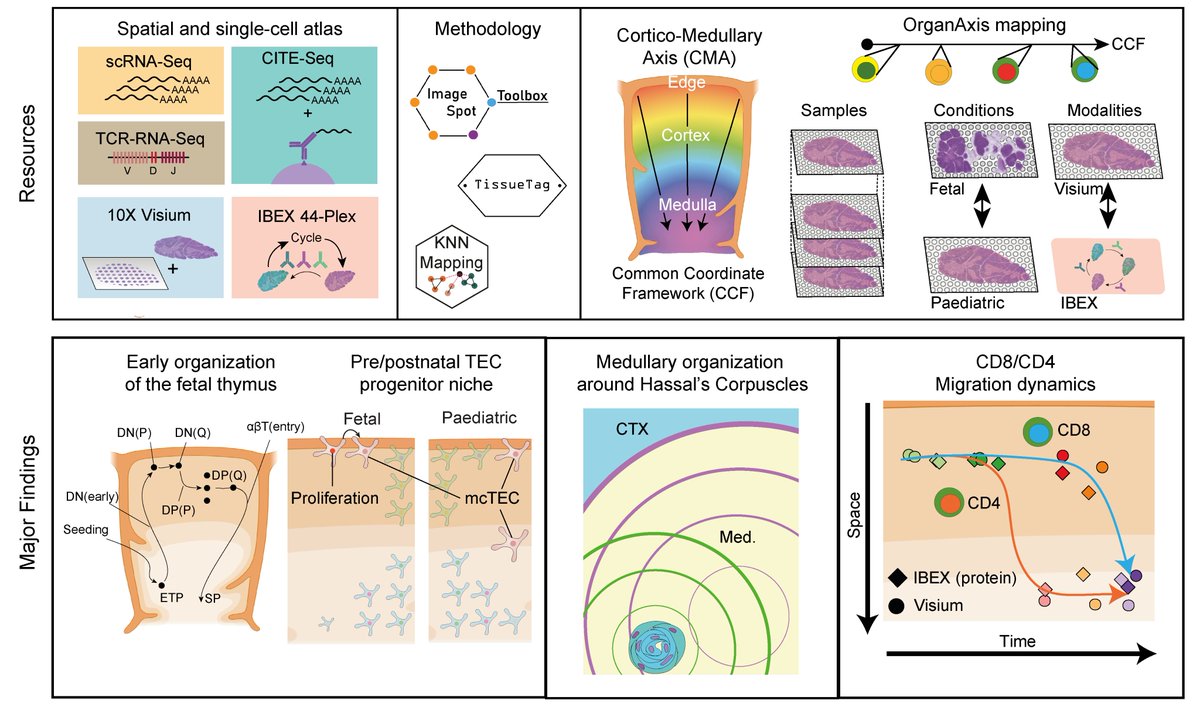
If you are into thymus, quirky TECs❤️, spatial mapping and CCF frameworks-check our preprint below!👇🚨 It was a huge pleasure to pull this off with our creative inventor Nadav Yayon🕵️♂️, T cell guru Lena Boehme👩🔬, imaging wizard Andrea Radtke🧝♀️ our labs and collaborators!!! 🤗🎉






Honoured to receive the Otto Schmeil prize by Heidelberger Akademie der Wissenschaften! Grateful for the recognition of our research on cerebellum development and evolution. Huge thanks to Henrik Kaessmann Lab and my closest colleagues Ioannis Sarr and Kevin Leiss!


Excited to share my PhD work in the lab of @larsplus Centre for Genomic Regulation (CRG). We screened the activity of synthetic enhancers throughout myeloid differentiation. Combinations of activating TFs frequently neutralized each other or gained repression twtr.to/wiWIZ (1/n)

We are excited to announce an opportunity for a Research Assistant position in our group Cambridge Stem Cell Institute Cambridge University to help with our work on the Human Cell Atlas using #organoids jobs.cam.ac.uk/job/48837/

It was a great honor to receive this award and a fantastic opportunity to return to Heidelberg and see many friends. I am extremely grateful for having had the opportunity to work with Kaessmann Lab and am already looking forward to my next visit! 🤗

![Ibrahim I. Taskiran (@ibrahimihsan) on Twitter photo Exciting news! Our latest research on "Cell type directed design of synthetic enhancers" has been published in <a href="/Nature/">nature</a>. We've harnessed deep learning to design enhancers with cell-type specificity. Find our paper here: nature.com/articles/s4158…. Highlights below: [1/n] Exciting news! Our latest research on "Cell type directed design of synthetic enhancers" has been published in <a href="/Nature/">nature</a>. We've harnessed deep learning to design enhancers with cell-type specificity. Find our paper here: nature.com/articles/s4158…. Highlights below: [1/n]](https://pbs.twimg.com/media/GBKA6x7XQAAn_7m.jpg)


![Niklas Kempynck (@niklaskemp) on Twitter photo Happy to share CREsted, our latest package on enhancer modeling! CREsted allows for training sequence-based deep learning models from scATAC-seq data, enhancer decoding and enhancer design through a variety of methods.
Check out the GitHub here: github.com/aertslab/CREst…. [1/9] Happy to share CREsted, our latest package on enhancer modeling! CREsted allows for training sequence-based deep learning models from scATAC-seq data, enhancer decoding and enhancer design through a variety of methods.
Check out the GitHub here: github.com/aertslab/CREst…. [1/9]](https://pbs.twimg.com/media/GV6688OXcAgK-iH.png)