CRI iAtlas
@iatlas_cri
Platform & analytic tools for exploring molecular and cellular response in cancer immunotherapy. Powered by @isbsci @SageBio @bgvincentlab @CancerResearch
ID: 1111643385427746817
https://www.cri-iatlas.org 29-03-2019 14:56:51
86 Tweet
107 Followers
19 Following







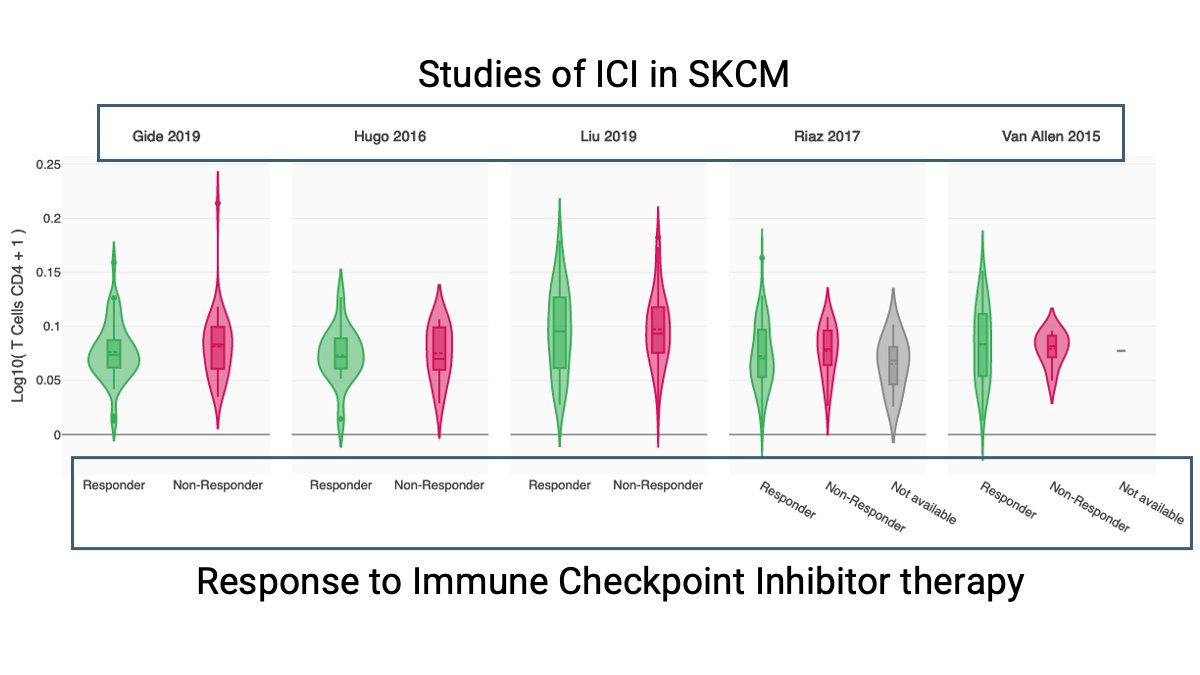
What is the CRI iAtlas CRI iAtlas, and how is this interactive tool accelerating discovery for a future #Immune2Cancer? CRI iAtlas Grantee Dr. James Eddy James Eddy Sage Bionetworks describes its value in exploring and analyzing immuno-oncology data during Cancer Immunotherapy Month.




New data available in CRI iAtlas! iAtlas now includes immunogenomic data derived from the PRINCE (pancreatic cancer) and PORTER (prostate cancer) immunotherapy trials. This update also includes data on breast cancer checkpoint treatment from @NCIHTAN. Cancer Research Institute

New data available in CRI iAtlas! iAtlas now includes immunogenomic data derived from the AMADEUS immunotherapy trial. rupress.org/jem/article/22… Cancer Research Institute

We've posted another Newsletter! Dive in to learn about AACR Project GENIE, a novel feature from a #GSoC participant, new data from CRI iAtlas & The Cancer Dependency Map, and more. The newsletter also has details on a cBioPortal symposium on Nov 11 in Amsterdam. buff.ly/7gnccvd