Harold D. Kim
@haroldkimlab
I'm a physics professor at Georgia Institute of Technology aka Georgia Tech or GT. #BiologicalPhysics #PhysicsofLivingSystems
ID: 3236308856
https://haroldkimlab.gatech.edu/ 04-06-2015 20:03:58
1,1K Tweet
594 Followers
411 Following


Do you need to quantify proteins in #cellfree gene expression systems but don't want to use a bulky fluorescent protein? Jessica Willi and Ashty Karim report the development and use of a fluorescent minihelix in ACS Synthetic Bio that can help pubs.acs.org/doi/full/10.10…

Journal cover Nucleic Acids Res highlights our new method, RNAscape, for mapping base pairing and tertiary interactions in RNA structure: academic.oup.com/nar/article/52…. Same NAR issue features our new deep learning method to predict DNA shape: academic.oup.com/nar/article/52… USC Dornsife College of Letters, Arts & Sciences USC Research





Scott C Blanchard Congratulations to all the authors




Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. Bruno Correia Sergey Ovchinnikov biorxiv.org/content/10.110…

“I had always had this impression that to do computer science, you had to be a hacker and know everything about Linux. I realized that theoretical computer science was as fascinating as theoretical physics, and I said, ‘OK — this is what I want to do.’” quantamagazine.org/the-computer-s…

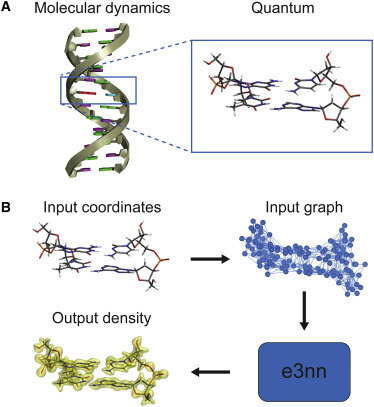
Predicting accurate ab initio DNA electron densities with equivariant neural networks by Alex J. Lee, Josh Rackers and William P. Bricker hubs.li/Q02YHY8c0 Biophysical Journal highlight — An official journal of Biophysical Society


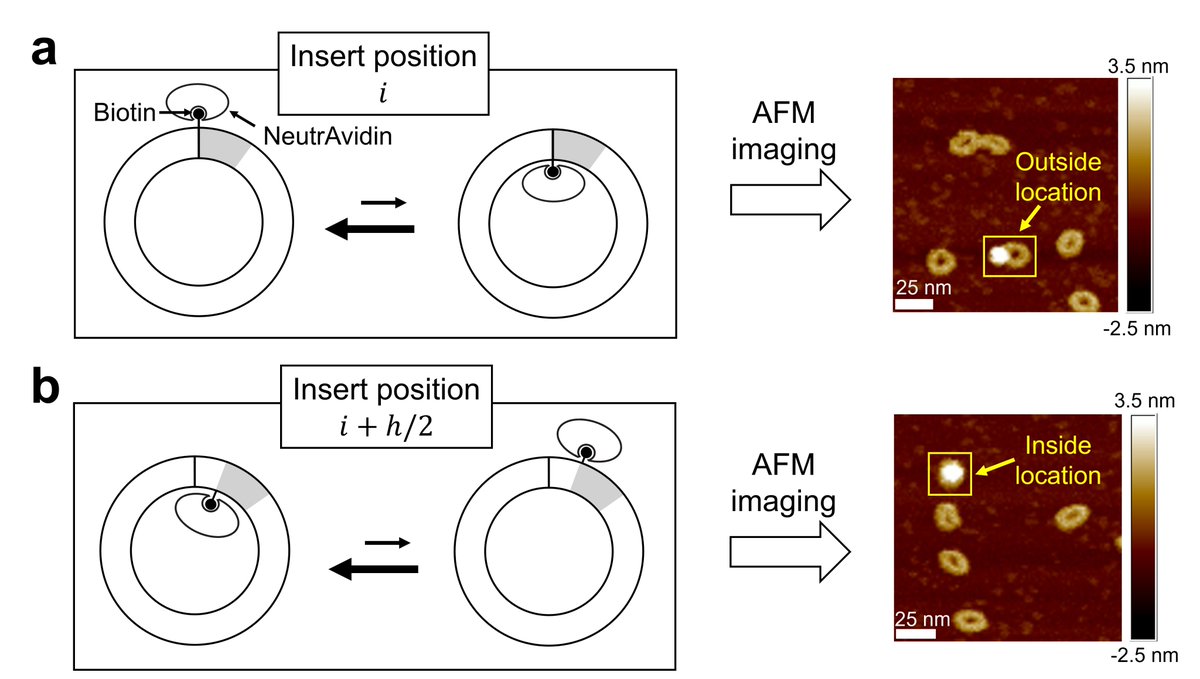
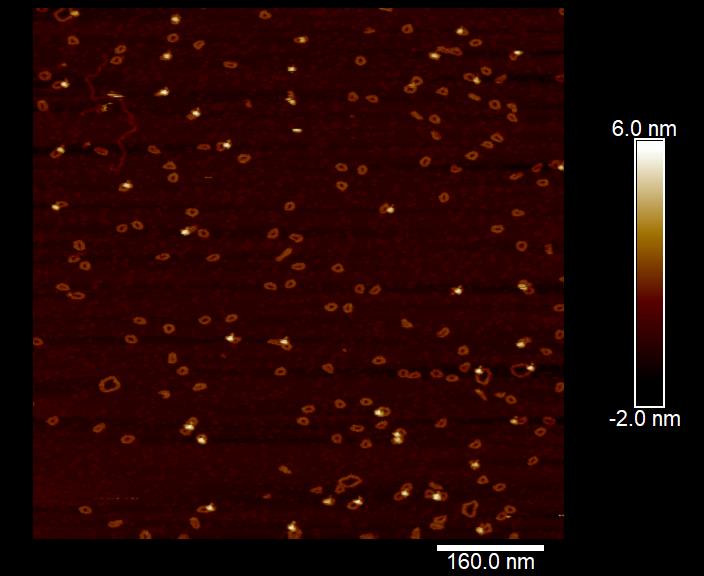
The Harold D. Kim studies #DNA molecules far too small to be resolved with an optical microscope. Here, Tony Lemos uses atomic force #microscopy to image DNA minicircles as they bind to individual proteins!


🚨Preprint alert🚨 We show experimentally that DNA minicircles adopt a favorite “inside-out” orientation depending on their sequence. Big thanks to Tony Lemos (Tony Lemos) for leading the work! biorxiv.org/content/10.110…