
Haowen Zhang
@haowen_zhang
CSE PhD@Georgia Tech. Opinions are my own.
ID: 863000841778733056
http://haowenz.github.io 12-05-2017 12:00:09
162 Tweet
144 Takipçi
138 Takip Edilen





Minimap2 v2.22 released with a short preprint associated with it, more formally describing recent improvements since v2.19. Thank Chirag Jain, Arang Rhie and all users for their invaluable feedback. arxiv.org/abs/2108.03515





Our newest work Chromap for fast chromatin profiling analysis is published on Nature Communications today! Chromap significantly reduces the pain of waiting in the era of deep sequencing and single-cell analysis. Haowen Zhang Xiaole Shirley Liu Heng Li nature.com/articles/s4146…

Kar-Tong Tan found Oxford Nanopore basecallers often systematically call telomere repeats (TTAGGG)n into other repeats with similar signal profiles. He tuned the bonito basecaller and developed a pipeline to greatly reduce such errors. Read the thread and our preprint for details.




The previous hifiasm paper described haplotype-resolved assembly with trio data. In this new paper, Haoyu Cheng achieves chromosome-long phased assembly without parents. Tested for human and non-human species. A single executable and one command line for the entire process.


1/3 We released Chromap v0.2.3 with the ultimate flexible barcode specification (hopefully) Haowen Zhang ! It now supports the complex non-consecutive barcode specification, a very useful feature for SPLiT-seq data, e.g.: github.com/haowenz/chroma…

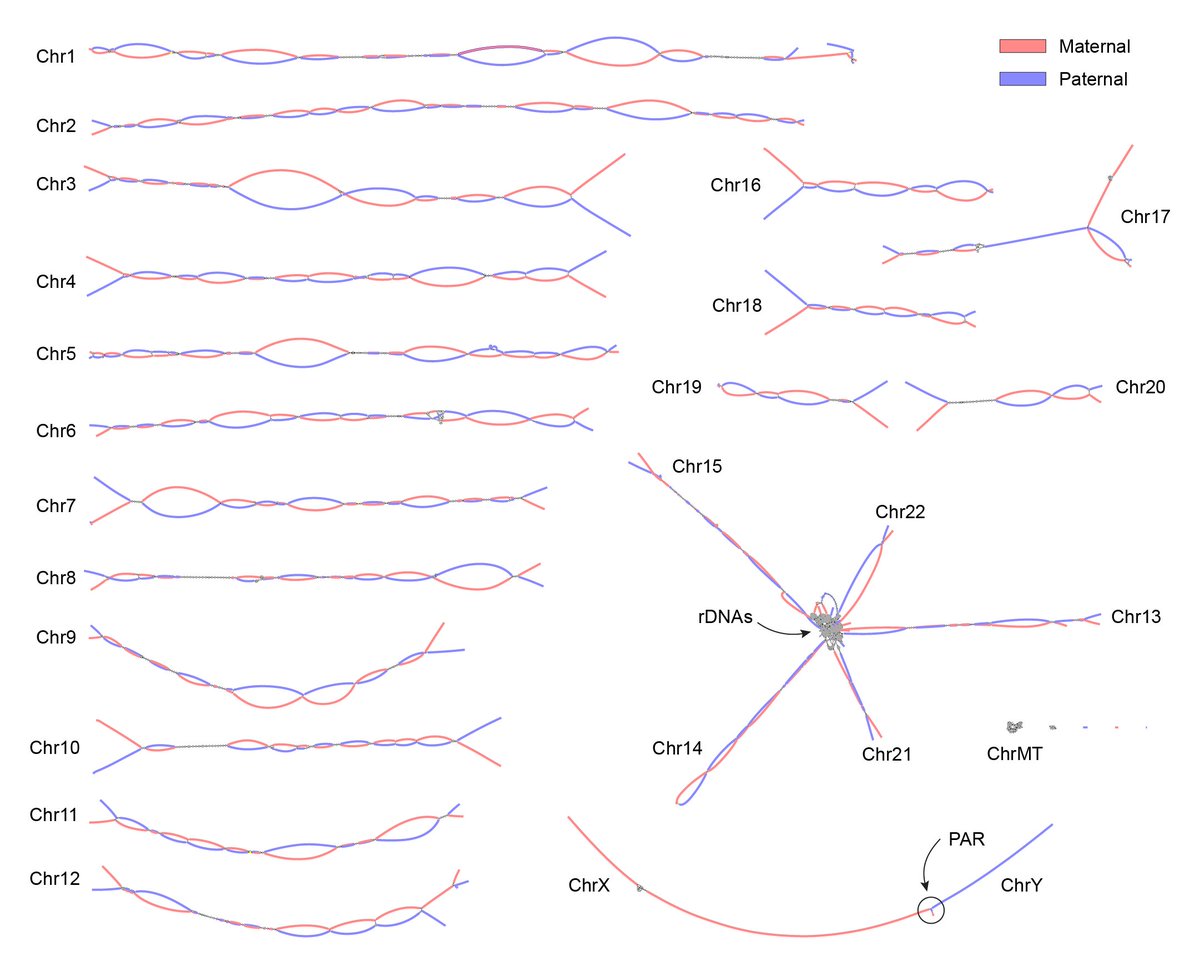
Our next big thing! 🎉 "Verkko: telomere-to-telomere assembly of diploid chromosomes" from Mikko Rautiainen Sergey Nurk 🇺🇦 Sergey Koren generates beautiful graphs and automatically assembles 20/46 human chromosomes from HG002 #T2T without gaps biorxiv.org/content/10.110…




![Chirag Jain (@chirgjain) on Twitter photo Most long-read assembly tools assume that the string graph model [Myers 2005] encodes every valid assembly, but this is unfortunately false. Excited to share new results on how to address this. Most long-read assembly tools assume that the string graph model [Myers 2005] encodes every valid assembly, but this is unfortunately false. Excited to share new results on how to address this.](https://pbs.twimg.com/media/FOQ2KVNaIAQTrPb.jpg)