Hamed Khakzad
@hamed_khakzad
Junior Professor @INRIA | PhD @UZH_en, postdoc @EPFL | Protein design | Mass Spectrometry | Deep Learning
ID: 1067459803369361408
27-11-2018 16:47:04
208 Tweet
286 Takipçi
270 Takip Edilen








Excited to be at #ICLR2025 and happy to chat about ML and drug discovery! 🇸🇬 Come find me at one of our posters or feel free to reach out directly! 1. DrugFlow: #9 Thu 24/04 10am 2. SynFlowNet: #15 Fri 25/04 10am 3. LDDM: GEMBio Workshop 27/04 4. LatentFrag: #AI4MAT 28/04





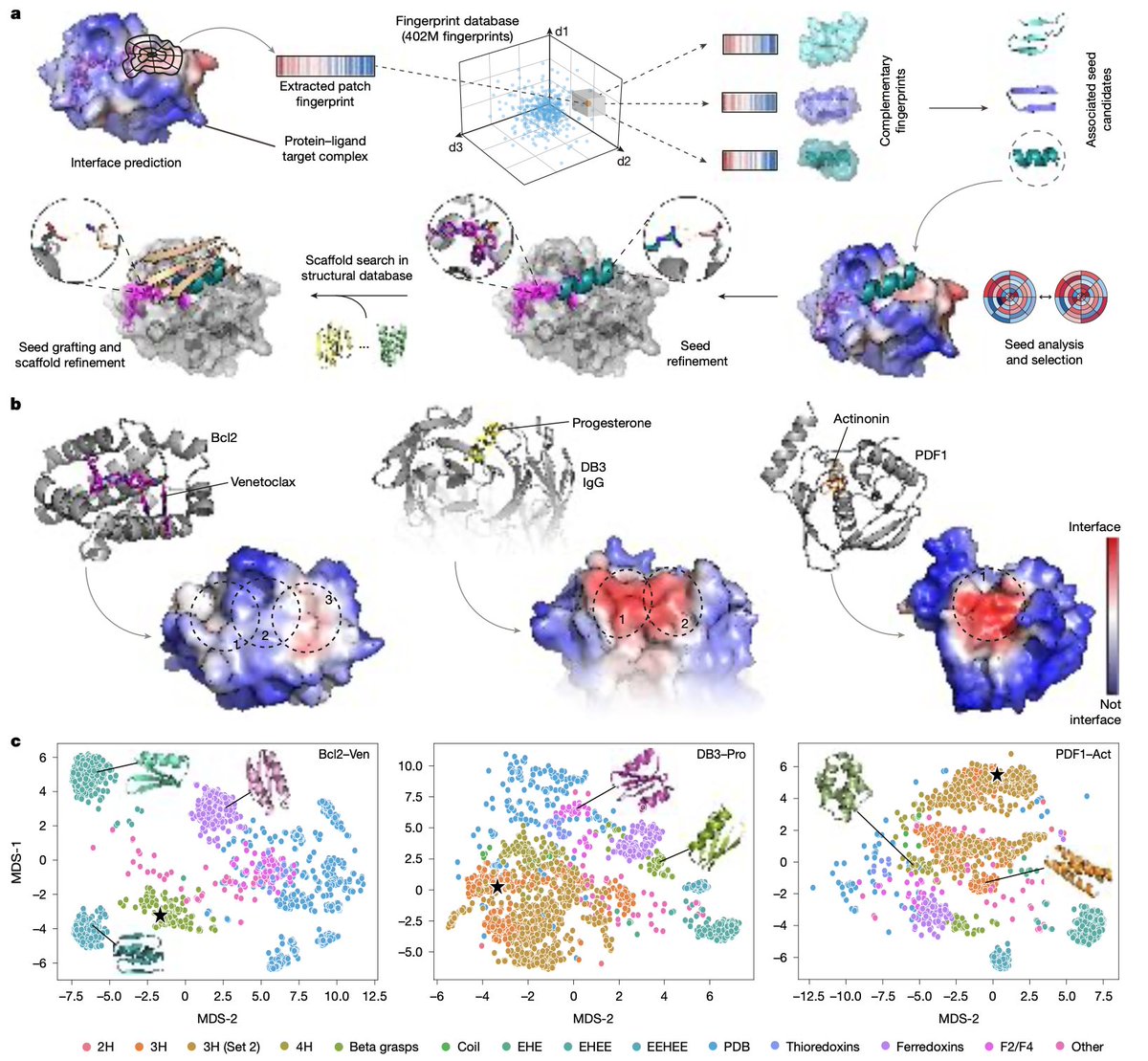
We were invited to write a preview about SHAPES, a great recent work from Possu Huang Lab. I really enjoyed this paper! It shows how far we still are from sampling the protein structural space without bias. Our preview just came out, check it out here: authors.elsevier.com/a/1ldzw8YyDfuZ…


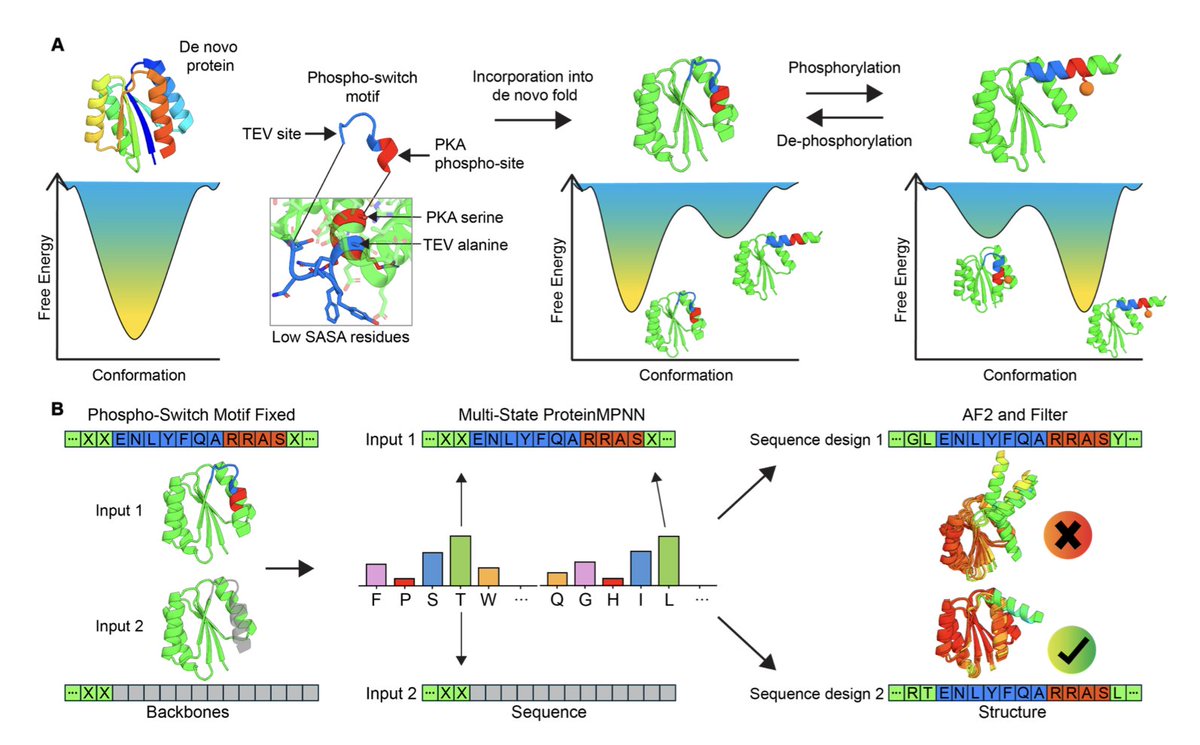
Exciting to see our protein binder design pipeline BindCraft published in its final form in nature ! This has been an amazing collaborative effort with Lennart, Christian Schellhaas, Sergey Ovchinnikov, Bruno Correia and many other amazing lab members and collaborators. nature.com/articles/s4158…