
Genki Terashi
@gterashi
Assistant Research Scientist @Purdue University My research projects: Protein structure modeling, prediction, Protein structure Modeling from cryoEM data.
ID: 1070023929434202112
04-12-2018 18:35:59
929 Tweet
225 Takipçi
147 Takip Edilen



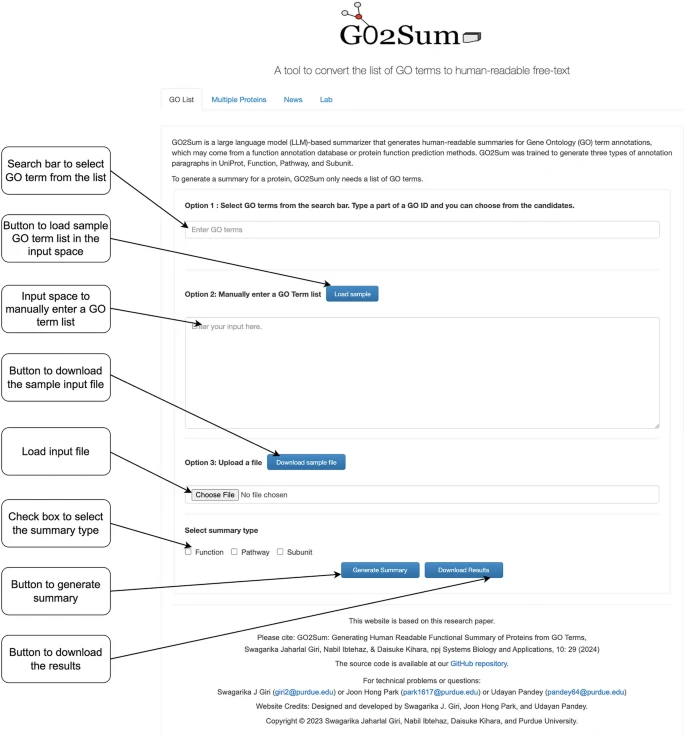
New book chapter online: Translating a GO Term List to Human Readable Function Description Using GO2Sum by Swagarika Jaharlal Giri Udayan Pandey, joonhongpark & Daisuke Kihara Methods in Mol. Biol. The server is available kiharalab.org/go2sum_server/ link.springer.com/protocol/10.10…



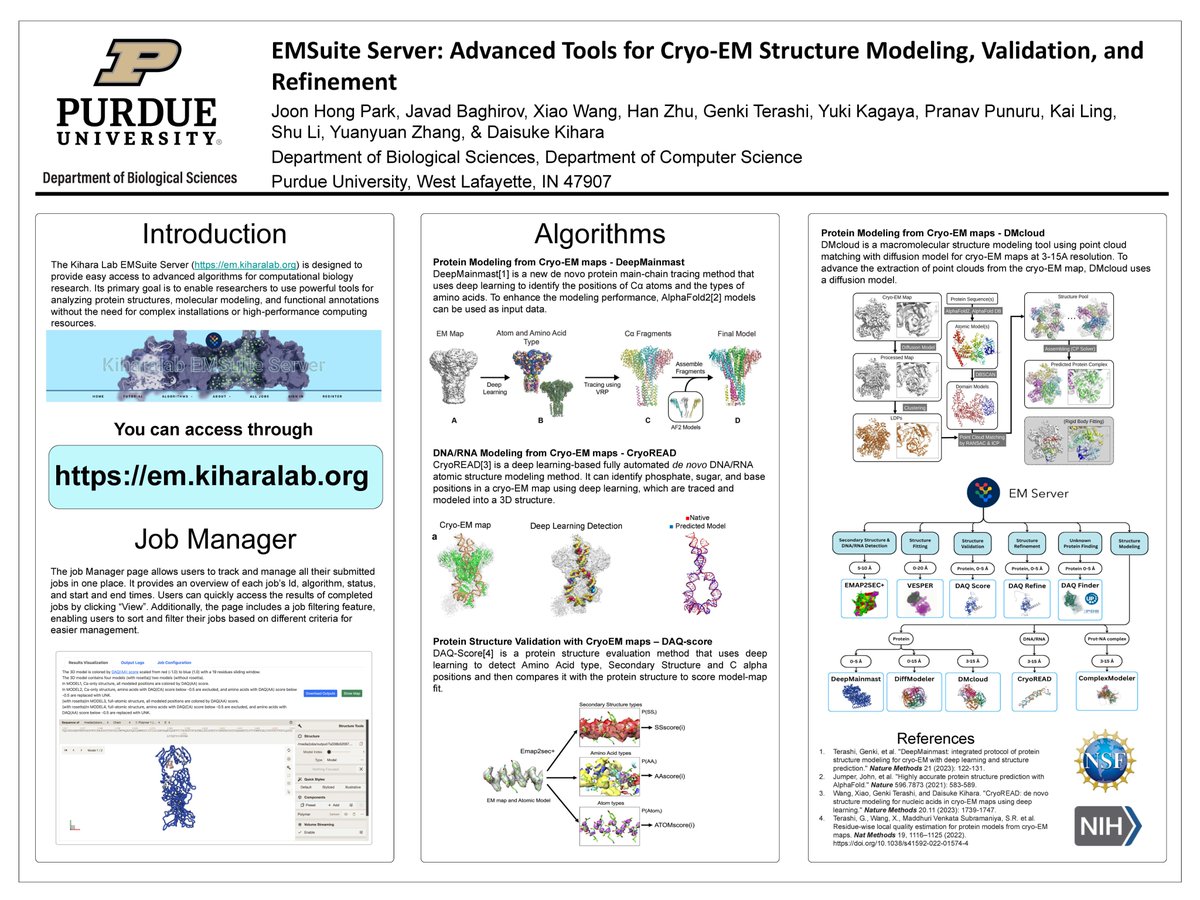
Joon Hong Park from Purdue Computer Science at American Crystallographic Association (ACA) 2025 meeting. presenting his poster about our cryo-EM structure modeling server. Visit the server for easy and accurate modeling at em.kiharalab.org


New paper accepted: "Structure Modeling Protocols for Protein Multimer and RNA in CASP16 with Enhanced MSAs, Model Ranking, and Deep Learning" Yuki Kagaya et al., Proteins. This reports our strategies in CASP. The initial submission available at ggnpreprints.authorea.com/doi/full/10.22…


David Platt summarizing his summer research with us at the REU undergraduate symposium. Great job on an awesome presentation! Purdue Structural and Comp. Biology & Biophysics Purdue Biological Sciences Purdue Science









Thank you for coming to the DAQ and DAQ-Refine cryo-EM structure validation workshop hosted by Purdue Structural Biology and Biophysics Club ! For those who missed it, we have Youtube video: youtube.com/watch?v=YRAT98… The presented tools are available at em.kiharalab.org


New paper released! "Distance-AF improves predicted protein structure models by AlphaFold2 with user-specified distance constraints" Yuanyuan Zhang, Zicong Zhang, Y Kagaya, G Terashi, B Zhao, Y Xiong & D Kihara, Communications Biology. Communications Biology nature.com/articles/s4200…







