Sergei Grudinin
@grudinin
Structural bioinformatician, group leader @inria_grenoble and @CnrsAlpes
ID: 55658845
https://team.inria.fr/nano-d/ 10-07-2009 20:43:25
34 Tweet
242 Takipçi
176 Takip Edilen

Mohammed AlQuraishi Connor W. Coley Thanks, our perspective (for the casp issue) can be found here arxiv.org/abs/2105.07407





Our work with @ilvailvail and Nikita Pavlichenko is published today! What a day for #AI and protein structure prediction :)

by popular demand, Sergey Ovchinnikov is going teach us how to use #RoseTTAFold and #AlphaFold2 on Wednesday, August 4th at 7pm EDT Sergey Ovchinnikov "ColabFold - Making protein folding accessible to all via Google Colab!" harvard.zoom.us/j/94214636314

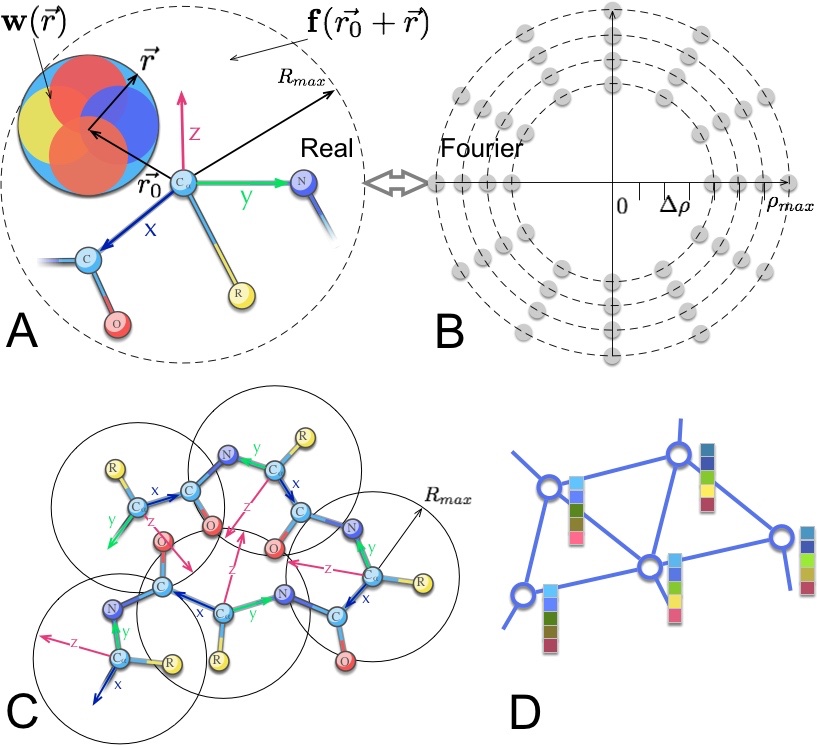
Proud to share the first work of Dmitrii Zhemchuzhnikov with assitance from @ilvailvail on our vision of 6DCNNs, roto-translational convolutions, local SE(3) equivariance and a mix of protein representations! arxiv.org/abs/2107.12078 #6dcnn #equivariance


What are the ingredients for protein partner identification? Check out our preprint reporting on a molecular cross-docking-based approach for the ab initio reconstruction of protein-protein interaction networks! Alessandra Carbone Yasser MOHSENI LCQB biorxiv.org/content/10.110…


Thematic School 2021 : Graph as models in life sciences: #Machinelearning and integrative approaches 📍Online ⏰25th – 29th of October 2021 َ Sergei Grudinin Jean Louis Raisaro @cazencott



PhD proposal to work on alternative splicing and protein design. Selected candidates will be funded for 4 years (pre-doc+PhD). Apply before March 24! ibio.sorbonne-universite.fr/wp-content/upl… ibio.sorbonne-universite.fr/2022-program/ i-Bio Sorbonne Université Sergei Grudinin

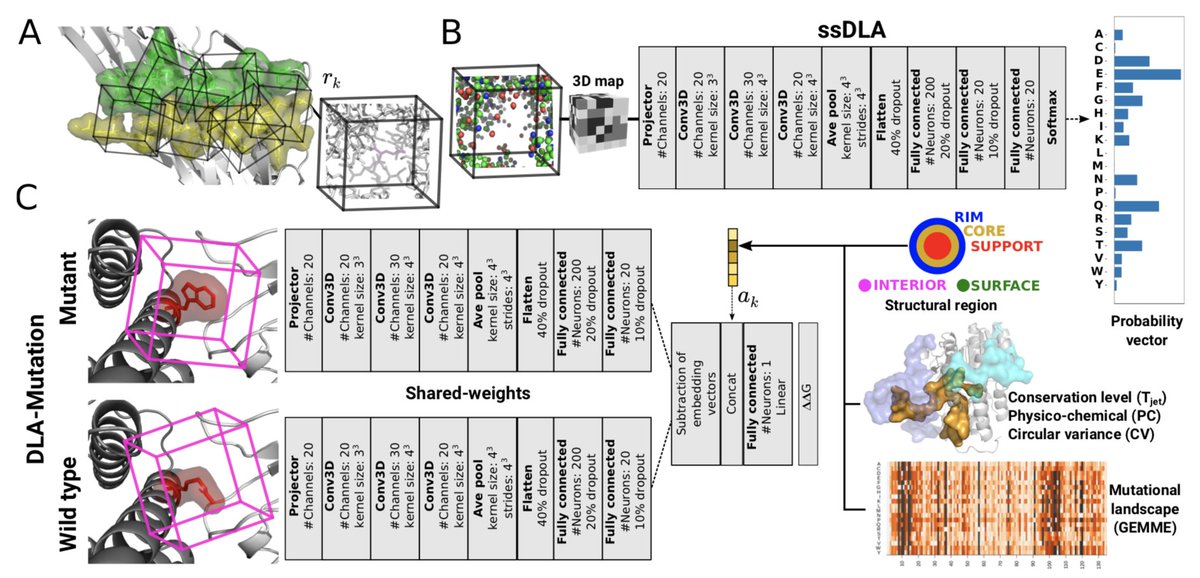
Very happy to expand our Deep Local Analysis series! Protein interfaces represented as many residue-centred locally oriented cubes. Step 1: what's in the cube? Step 2: what's the effect of mutating that residue? thanks to Yasser MOHSENI Alessandra Carbone for the great collaboration!

Protein sequences to motions, great PhD work by Valentin Lombard, with Sergei Grudinin! We investigated whether compact continuous representations of protein motions can be predicted directly from sequences, without relying on or sampling protein structures. biorxiv.org/content/10.110… 1/3