The Gilbert Lab
@gilbertlabrna
We love RNA
ID: 1040274543523364864
13-09-2018 16:22:33
111 Tweet
2,2K Followers
51 Following

Don’t miss Rachel Niederer sharing her work identifying novel translational control elements during the mechanisms and regulation of translation session this morning at #RNA2021!


It's out! Dr. Yu Sun in the lab identified merafloxacin as a pan-betacoronavirus programmed -1 ribosomal frameshift inhibitor. Great collaboration with 𝙱𝚛𝚎𝚝𝚝 𝙻𝚒𝚗𝚍𝚎𝚗𝚋𝚊𝚌𝚑 🌊 Wendy Gilbert Rachel Niederer Craig Wilen Ya-Chi Ho and YCMD colleagues Yale West Campus pnas.org/content/118/26…


Huge congratulations to Nicole M Martinez who is starting her own lab Stanford University! Keep an eye out for exciting work on #RNA processing and modifications!




Huge congratulations to Cole Lewis for getting the NOA on his F31 this week!!! #sweet16 🎊🍾🎉🎊🍾🎉


Excited that the Martinez Lab Stanford Medicine Stanford Chemical and Systems Biology Sarafan ChEM-H Dev Bio @ Stanford is hiring! We are looking for a Research Assistant to work on projects related to how RNA modifications and mRNA processing control gene expression. Apply - job ad here: careersearch.stanford.edu/jobs/life-scie…

Check out the latest work from our lab! Congratulations to Rachel Niederer @bzinsh and Mariarojas! 🎉🎉🎉 authors.elsevier.com/c/1eR4-8YyDffJ…

More good news! 🎉 Here we uncover roles for pseudouridylation and pseudouridine syntheses in alternative mRNA processing. Spearheaded by the fantastic Nicole M Martinez, this was a collaborative effort with Gene Yeo’s Lab at UC San Diego Congratulations to all authors! sciencedirect.com/science/articl…

In a span of < 1 wk, The Gilbert Lab publishes 2 incredible ACS Research stories-1st w insights into how mRNAs are born, tumors & mets; & then American Cancer Society PF Rachel Niederer publishes cell.com/cell-systems/f… showing how these mRNA instructions are used, & how to manipulate them🤯

Congratulations Rachel Niederer!! We can’t wait to see what cool new discoveries you make about translational control! 🎉

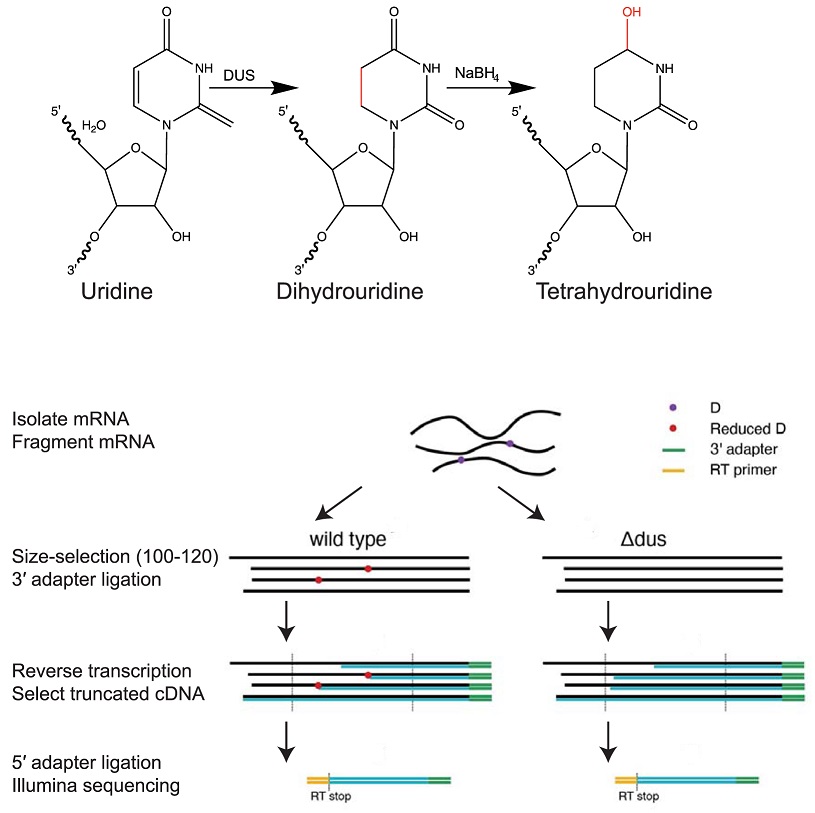
Novel method (D-seq) for transcriptome-wide high-res mapping of dihydrouridine reveals this #RNAmodification at new locations across the yeast transcriptome, suggesting broad role in folding functional RNA structures Austin Draycott The Gilbert Lab #PLOSBiology plos.io/3wR5Mta


Congratulations to our rock star grad students Cassandra and Cole Lewis who both won poster awards at #RNA22! 🎉🎉🎉

