Fodor Laboratory
@fodorlab
We're a molecular and structural virology lab working mainly on influenza @Dunn_School, @UniofOxford
ID: 1329367789501624322
https://fodor.path.ox.ac.uk/ 19-11-2020 10:16:02
313 Tweet
1,1K Followers
460 Following


Very pleased to have been able to present my work at my first Options XII for the Control of Influenza conference in Brisbane, Australia! It’s been an amazing time of meeting old and new colleagues and discussing incredible science 🦠 spot the koala 😉🐨 #influenza Options2024



Very proud of our latest preprint - using single-molecule assays to explore SARS-CoV-2 replication and inhibition. Thanks to all awesome co-authors Dani Groves Andrew McMahon Haitian Fan Jeremy Keown Fodor Laboratory Read it here: doi.org/10.1101/2024.1…


There is nothing more scary than a pumpkinfluenza! Rescued by Fodor Laboratory graduate student not-on-X Tom Shimkus.


The abstract submission for the Annual Meeting of the Society for Virology (GfV, GfV - Gesellschaft für Virologie) is open till 1 December 2024! GfV is officially affiliated with our Journal of General Virology (#JGenVirol).Don't miss the opportunity to share your research! microb.io/3NMmClQ


An image representing our work made the cover of Biophysical Reports! If you're interested check out the paper at cell.com/biophysreports…. Thanks again to everyone involved Hutchinson Lab Swetha Vijayakrishnan Dani Groves Hafez El Sayyed and not-on-X Andrew McMahon :)




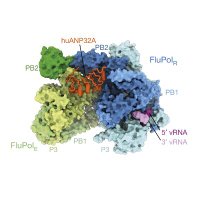
What's going on with bird flu in the UK at the moment? I wrote a piece for The Conversation explaining what we know at the moment, and why it's not (yet) time to worry theconversation.com/bird-flu-cases…

Virologist Wendy Barclay: ‘Wild avian viruses are mixing up their genetics all the time. It’s like viral sex on steroids’ Always worth listening to wendybarclay Imperial College London theguardian.com/science/2025/f…


Decoding viruses. Defying limits. 🦠🔍 Prof wendybarclay has dedicated her career to unpicking viral mysteries. Now, she's been appointed Regius Professor—the first woman to hold the prestigious title at Imperial College London ⤵️ #WomenInSTEM imperial.ac.uk/Stories/Wendy-…


Europe needs a sustainably funded influenza research and response network - especially in the light of H5N1 and current developments in the US. Read our commentary in The Lancet ID: thelancet.com/journals/lanin… With wendybarclay, Marion Koopmans, publications: https://pure.eur.nl and many other colleagues.

📢 Exciting job opportunities!📢 Two postdoc positions are available in our lab at Dunn_School (now @DunnSchool.bsky.social) to study influenza virus transcription, genome replication, nuclear export and assembly. More info: my.corehr.com/pls/uoxrecruit…



🚨🔎 Out now in PLOS Pathogens! 🔎🚨 This project has taken us MRC-Uni of Glasgow Centre for Virus Research UofG MVLS on a wild and wonderful journey. I am excited to share our story of how influenza A viruses (IAVs) exploit cell death to facilitate its covert spread. N/1 journals.plos.org/plospathogens/…
