Fan Cao
@fancaoerik
PhD student at #LinderstrømLang, University of Copenhagen. Molecular dynamics simulation, integrative structural biology, biophysics and tennis.
ID: 1559817710510690304
17-08-2022 08:21:46
64 Tweet
41 Followers
109 Following







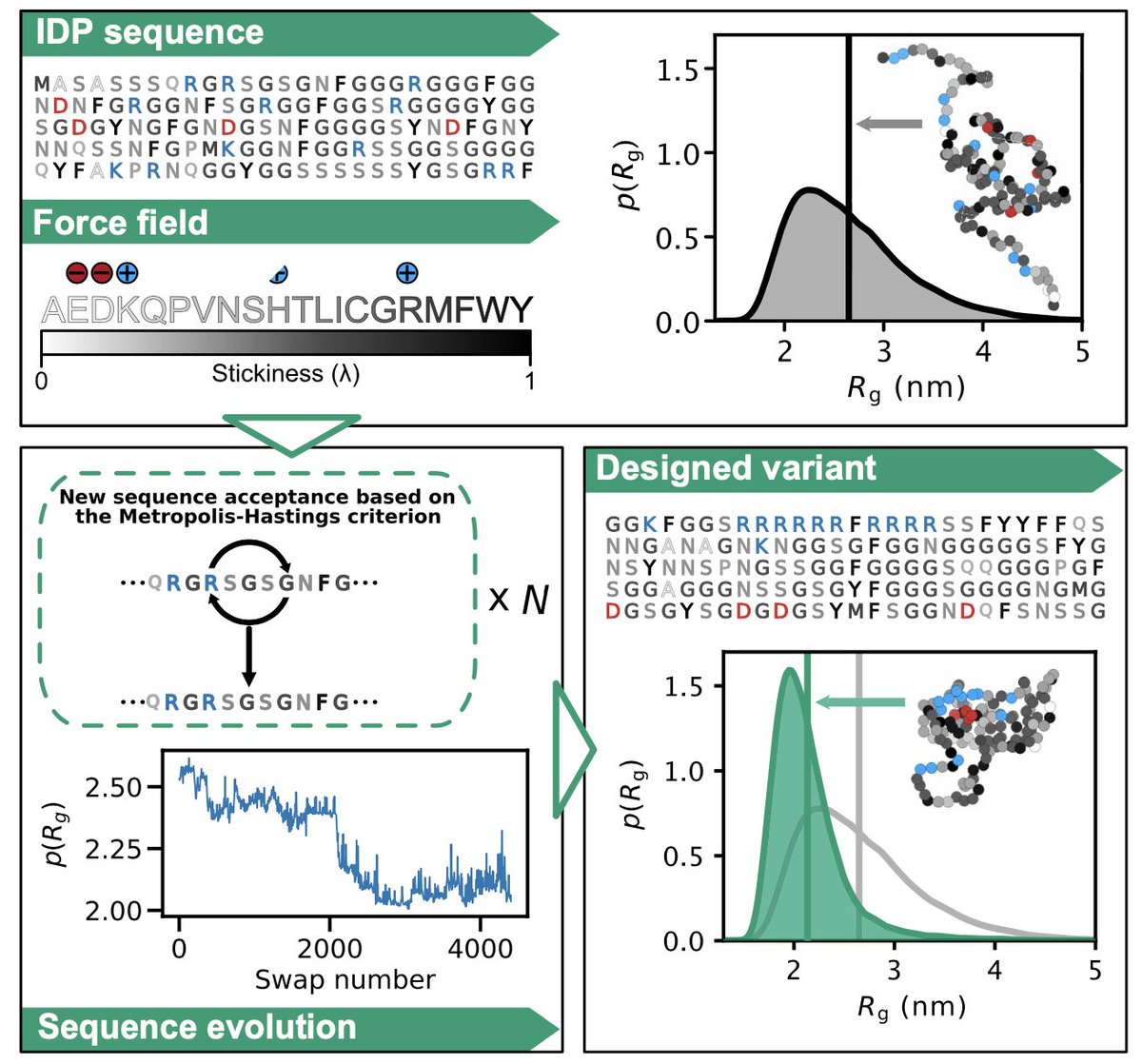
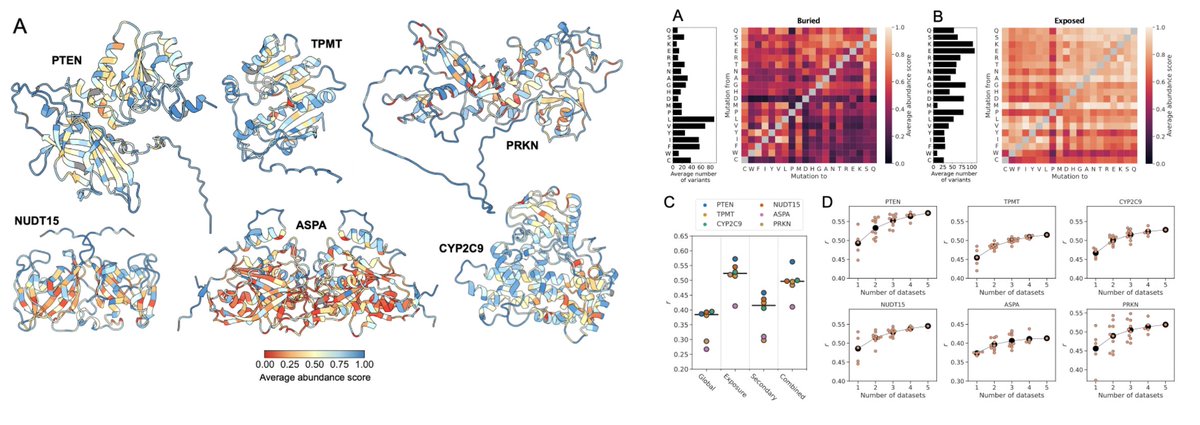
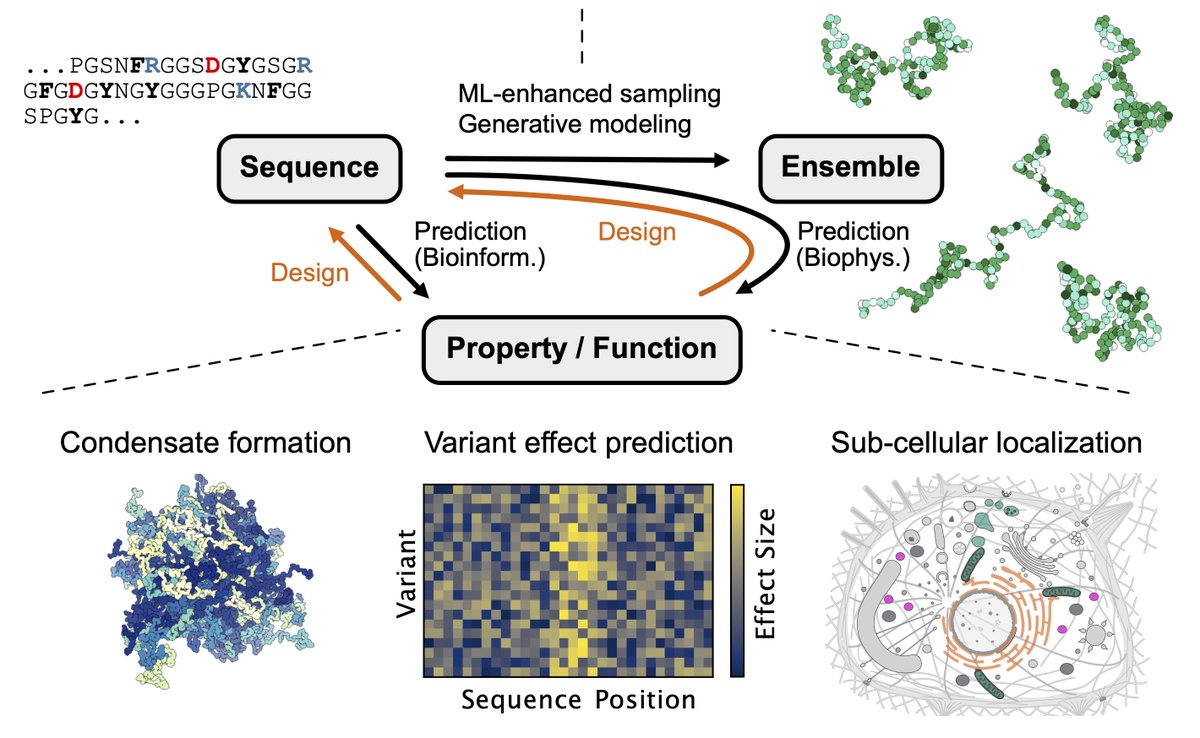
A great collaboration with KerstinDoerner and Maria Hondele! Also many thanks for the help from Kresten Lindorff-Larsen. We used our CALVADOS model to study the effect of tagging on condensation and phase separation—a good starting point for studying specific interactions.

Our paper on the use of dioxane as a reference for NMR diffusion measurements is now out authors.elsevier.com/a/1jxWn1SPT7tPu This paper led by Emil Tranchant revisits pioneering 25 y old work on using dioxane as a reference. We update the reference value and discuss the implications 1/4







Check out Rasmus Krogh Norrild's work with Alex Buell @biophysics_dtu and Joseph Rogers developing and using Condensate Partitioning by mRNA-Display to probe phase separation of ~100.000 sequences, and @SoBuelow simulations to support and analyse the experiments