Etienne Sollier
@etiennesollier
PhD student in Bioinformatics @DKFZ. Interested in cancer evolution, complex genomic rearrangements, epigenomics and AML.
ID: 1494968496190627844
19-02-2022 09:34:07
52 Tweet
222 Takipçi
174 Takip Edilen

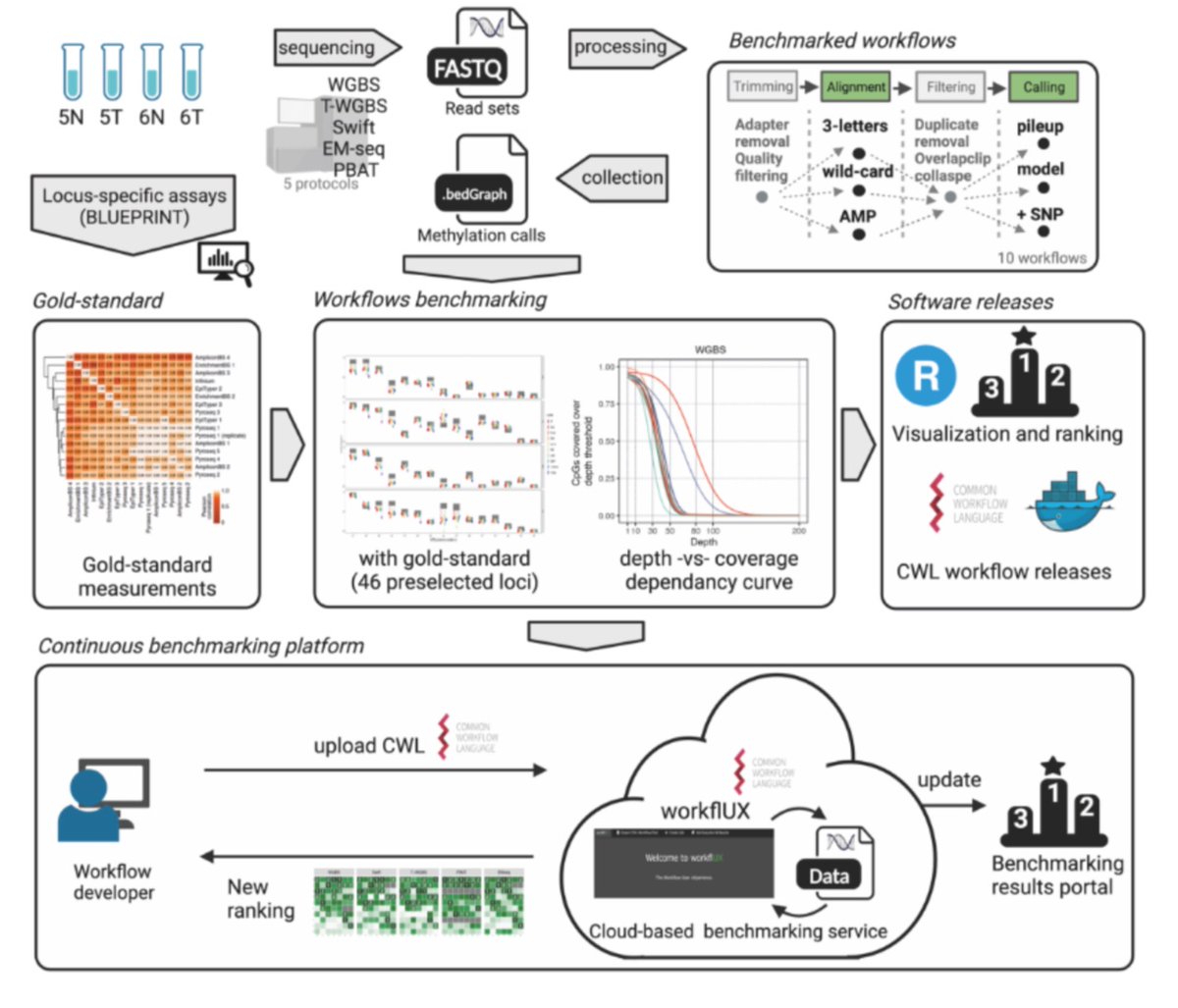
⛴️ On my way to Turku, Finland 🇫🇮, for #ECCB2024 ECCB2026 Geneva, Switzerland. If you want to learn about figeno, my visualization too for genomic data 🎨🧬 (github.com/CompEpigen/fig…), come to my talk at #ESCS2024 ESCS2024 tomorrow at 11:15, and to my poster (P15) on Wednesday at 6:30pm!


My poster for figeno (P15) is up at #ECCB2024 and I will present it this evening! Check it out if you want to visualize Oxford Nanopore, HiC, bigwig or WGS data! If you have already used figeno, I'd love to hear your feedback and suggestions for improvements. github.com/CompEpigen/fig…





An article published in Genome Biology presents epiCHAOS: a quantitative metric of cell-to-cell heterogeneity, applicable to any single-cell epigenomics data type. genomebiology.biomedcentral.com/articles/10.11…





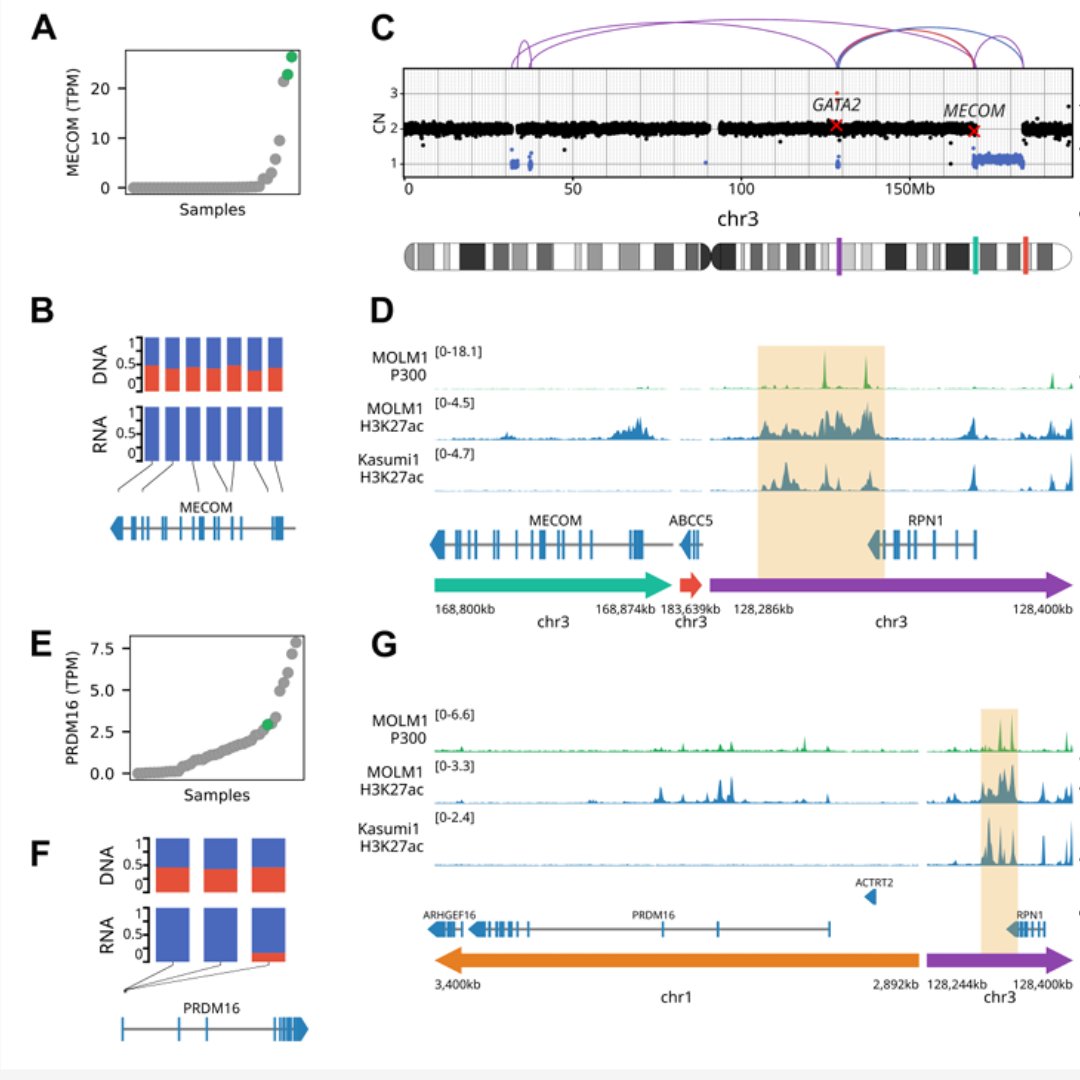
Our work on aberrant MNX1 expression in AML caused by enhancer hijacking is finally out! Great collaboration between the Plass lab, Translational Cancer Epigenomics and many others! Thanks a lot to everyone involved! 🙋🏻♂️