
Erik Sonnhammer
@eriksonnhammer
Professor of Bioinformatics at Stockholm University, Science for Life Laboratory. Network and Systems Biology; Orthology Inference; Protein Domains & Function.
ID: 795604493161340928
http://sonnhammer.org 07-11-2016 12:31:07
122 Tweet
273 Takipçi
55 Takip Edilen






Vårt projekt "Spatially and temporally resolved gene regulation networks to understand liver cancer formation" var ett av de 224 som fick anslag från Cancerfonden! Tack alla som skänker gåvor till forskningen och gör detta möjligt! #tillsammansmotcancer #cancerfonden

PhD position in #Generegulatorynetwork inference from spatial transcriptomics to understand regulatory mechanisms that lead to cancer formation. Location: SciLifeLab in Stockholm, Sweden. #bioinformatics #ComputationalBiology For details, see sonnhammer.org/download/ads/P…



Quest for Orthologs in the Era of Biodiversity Genomics academic.oup.com/gbe/advance-ar… GBE (Genome Biology and Evolution)


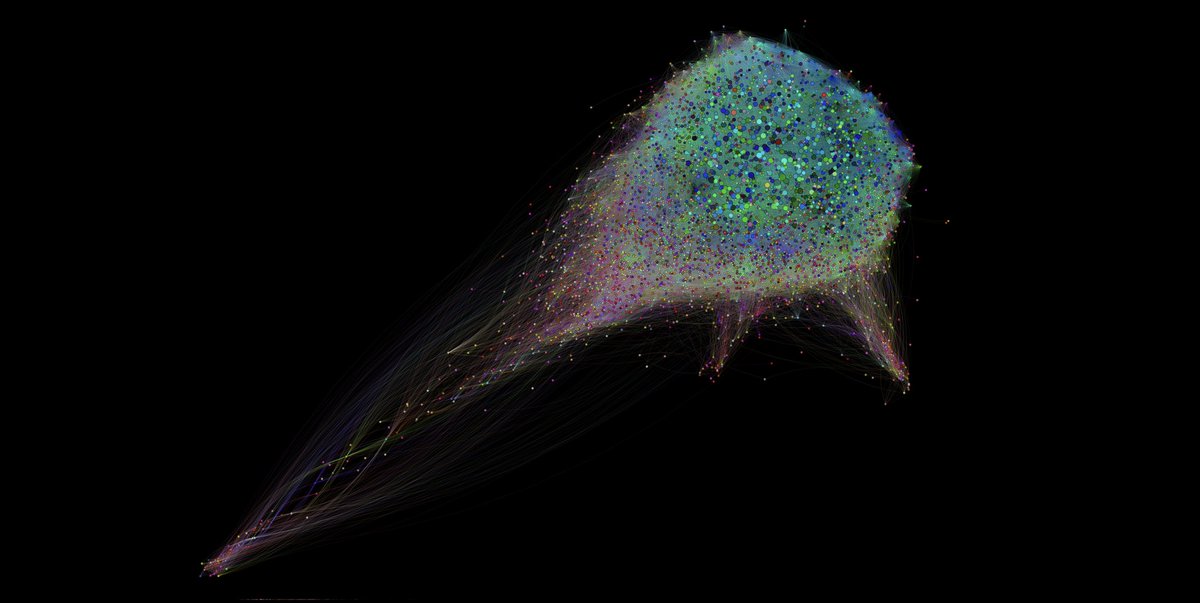
The human interactome represented by the FunCoup 5 network, illustrating interactions between 18,081 proteins connected by 5,036,826 links. By Davide Buzzao, Erik Sonnhammer, E. Persson, D. Morgan Enjoy the beauty of science? Check out our series #SciLifeLabArtTuesday on Instagram!


We are pleased to announce the publication of a new Pfam paper: The Pfam protein families database: embracing AI/ML Nucleic Acids Res academic.oup.com/nar/advance-ar…






PhD position open in #Generegulatorynetwork inference to develop new AI-based methods to improve simulations and accuracy. Location: SciLifeLab in Stockholm, Sweden. #bioinformatics #ComputationalBiology For details, see sonnhammer.org/download/ads/o…







