
Jungyoon Lee
@yololulu_
Master's @ Mila & Ex-finance Girlie @ JPMorgan | Generative Models, AI4Science, ML for chemistry
ID: 1496813760123650053
https://www.linkedin.com/in/jungyoon-lee-6a9aab139/ 24-02-2022 11:46:39
32 Tweet
78 Followers
201 Following






New work on scaling Boltzmann generators!!!! We went back to the basics for this one: 1.) no explicit equivariance 2.) more scalable transformer based flows than flow matching!!! Work co-led with charliebtan

SuperDiff goes super big! - Spotlight at #ICLR2025!🥳 - Stable Diffusion XL pipeline on HuggingFace huggingface.co/superdiff/supe… made by Viktor Ohanesian - New results for molecules in camera-ready arxiv.org/abs/2412.17762 Let's celebrate with a prompt guessing game in the thread👇



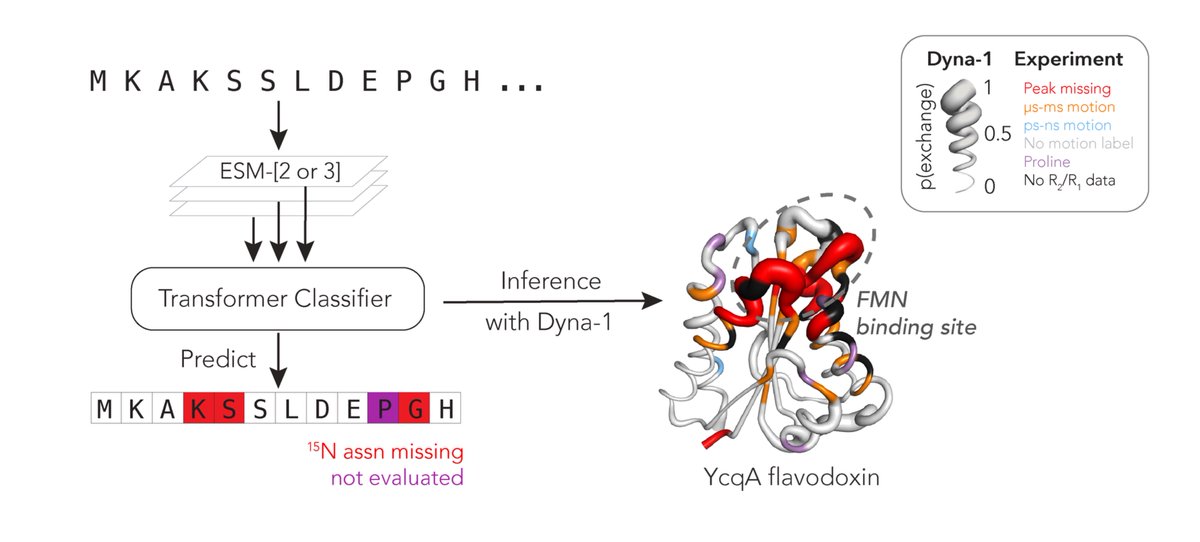
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵


(Hall 3 + Hall 2B #141 @ 3 pm) tomorrow OR infinity pool today! Super proud of this work by Marta Skreta AND Lazar Atanackovic AND Joey Bose AND Alex Tong AND me (I'm taking the photo (no)) 🥳


🧵(1/6) Delighted to share our ICML Conference 2025 spotlight paper: the Feynman-Kac Correctors (FKCs) in Diffusion Picture this: it’s inference time and we want to generate new samples from our diffusion model. But we don’t want to just copy the training data – we may want to sample

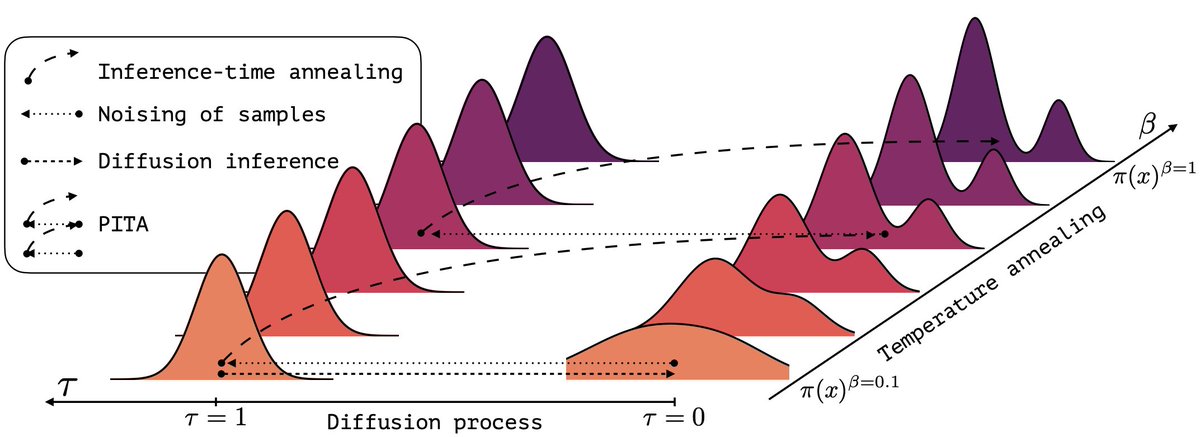
(1/n) Sampling from the Boltzmann density better than Molecular Dynamics (MD)? It is possible with PITA 🫓 Progressive Inference Time Annealing! A spotlight GenBio Workshop @ ICML25 of ICML Conference 2025! PITA learns from "hot," easy-to-explore molecular states 🔥 and then cleverly "cools"


🎉Personal update: I'm thrilled to announce that I'm joining Imperial College London Imperial College London as an Assistant Professor of Computing Imperial Computing starting January 2026. My future lab and I will continue to work on building better Generative Models 🤖, the hardest






