Tony
@tony_chen6
PhD Candidate @HarvardBiostats
ID: 1244061700342636547
29-03-2020 00:40:06
30 Tweet
98 Followers
249 Following



Excited to have you Aoxing Liu , really looking forward to your talk!!

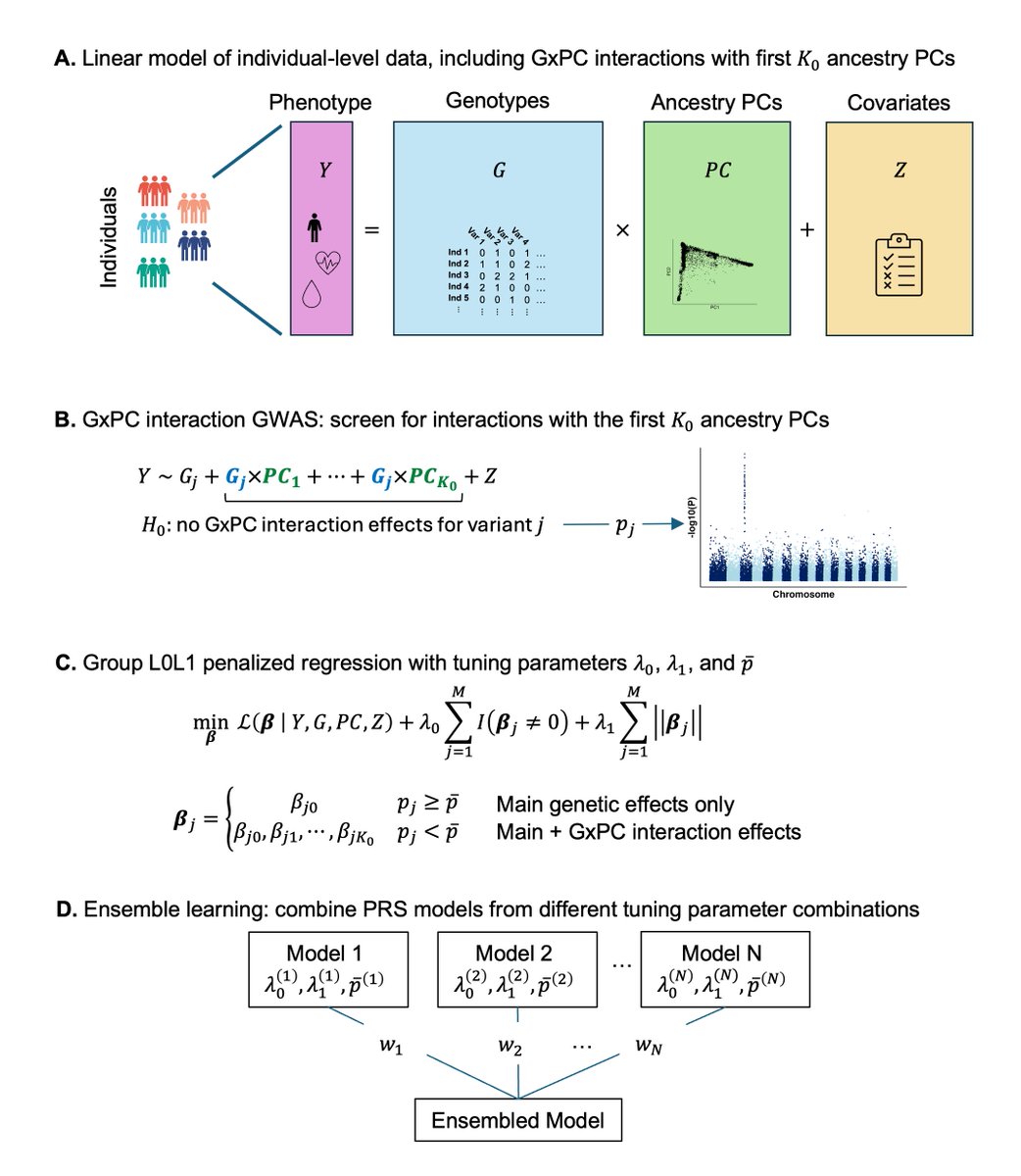
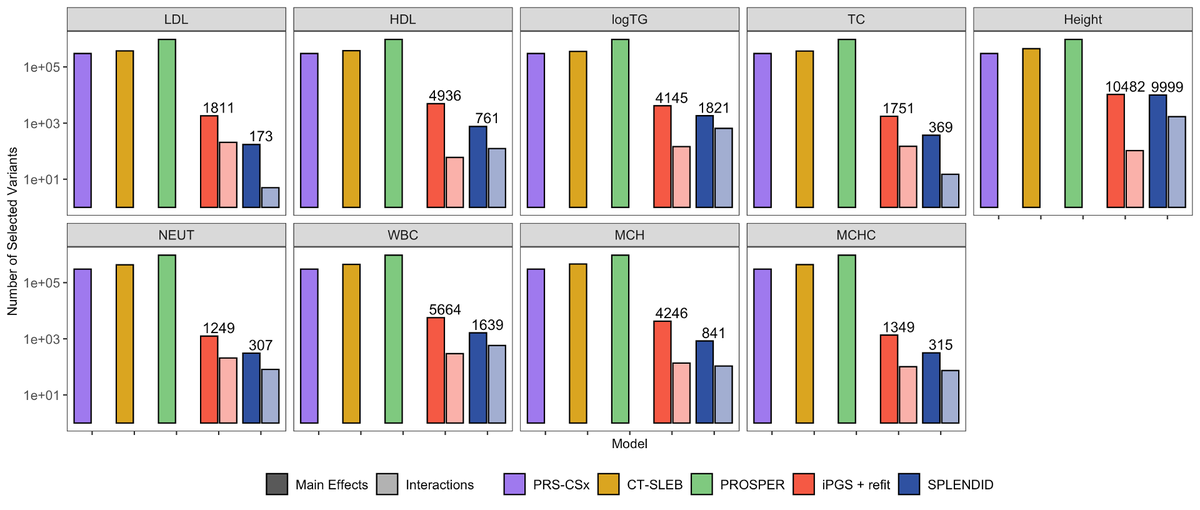
Excited to share our preprint for SPLENDID, a new PRS method using continuous ancestry interactions and biobank-scale individual level data. Many thanks to Haoyu Zhang , Rahul Mazumder, and Xihong Lin for their valuable guidance on this work! biorxiv.org/content/10.110…





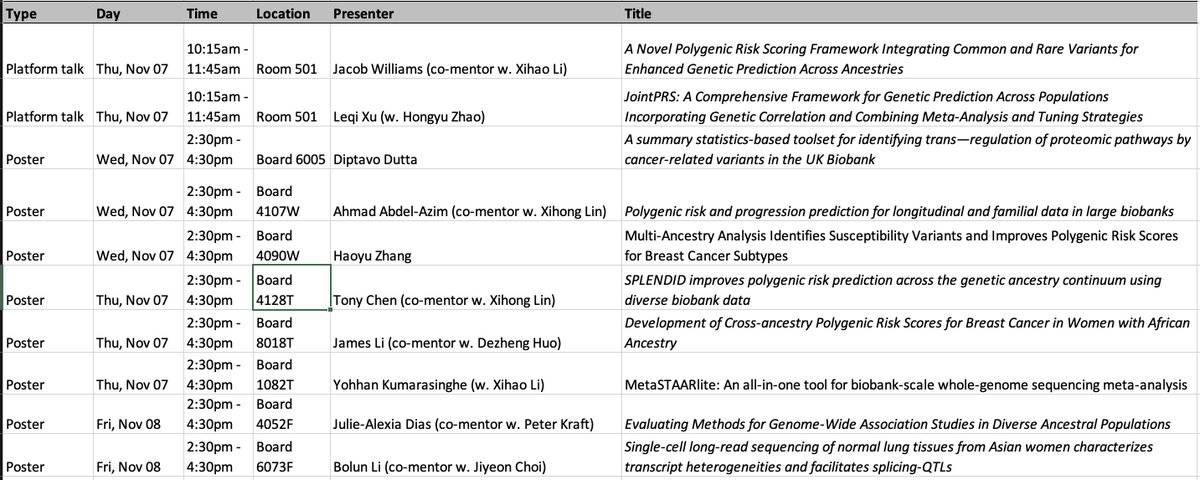
Check out the presentations from our lab and our close collaborators #ASHG2024. Such a hard-working year. So proud of the group!!! Very humbled to be able to work with so many great collaborators Xihao Li Xihong Lin Pete Kraft Diptavo Dutta Hongyu Zhao Tony Julie Alexia


Our paper introducing a PRS-based translation roadmap for lung cancer care is now published on eBioMedicine – The Lancet Discovery Science ! Wonderful collaboration with Haoyu Zhang , Li-Shiun Chen , ILCCO and GSCAN towards the clinical implementation of PRS. sciencedirect.com/science/articl…


Just in time! Come join us with Saori Sakaue on 11/19 for her recent work revealing genetic regulatory mechanisms through eQTL causal variants! Harvard Biostatistics Harvard Epidemiology


Please join us for our last HarvardPQG Working Group seminar of 2024, next Tuesday with Yosuke Tanigawa presenting his exciting recent work on polygenic prediction!

📢Out now! Xihao Li, Pradeep Natarajan, Zilin Li, Zhonghua Liu, Xihong Lin, and colleagues from NIH NHLBI present a framework for functionally-informed multi-trait rare variant analysis of biobank-scale sequencing studies. nature.com/articles/s4358… 🔓rdcu.be/d82Fx

Please join us tomorrow at HSPH for a HarvardPQG Working Group seminar with Kun-Hsing Yu , who will be discussing AI foundation models in pathology!


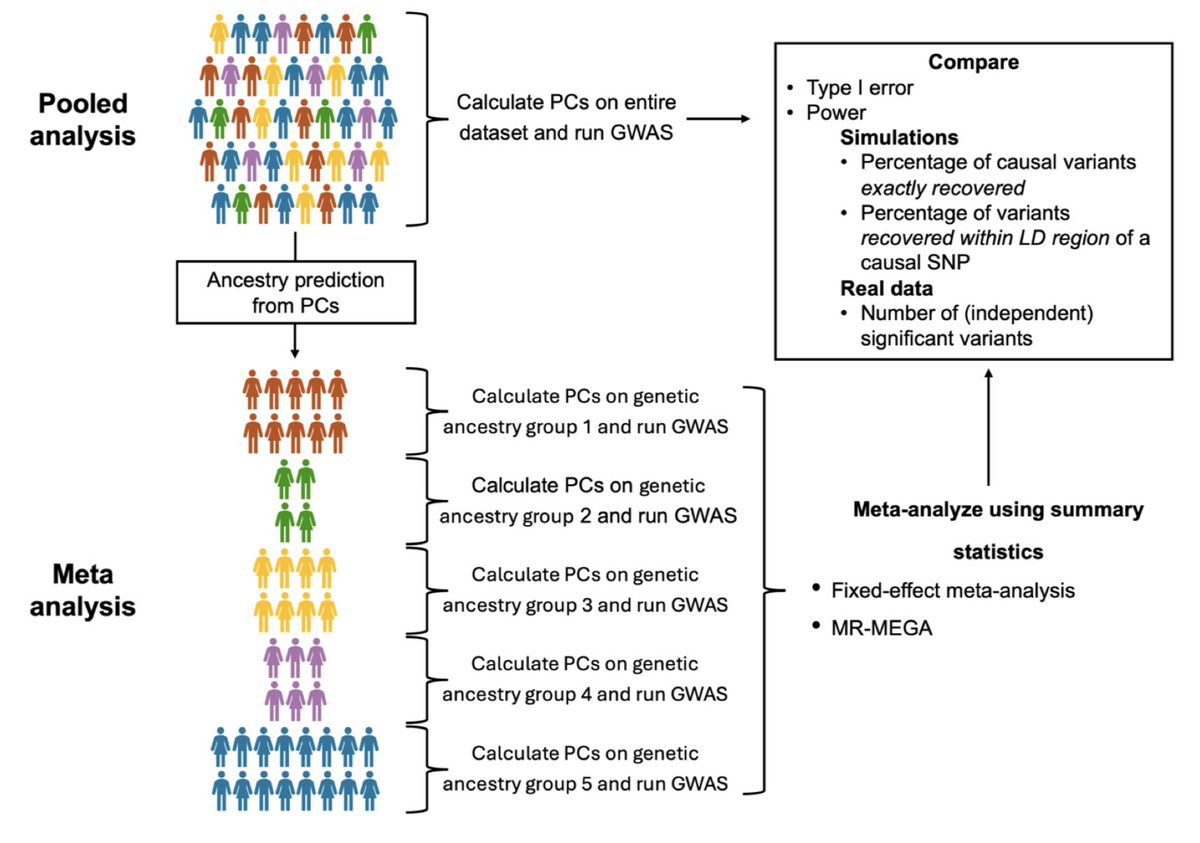
Excited to share our new manuscript on evaluating multi-ancestry GWAS methods using large-scale simulations & real data from UK Biobank (N≈324K) & All of Us (N≈207K)! My first thesis paper under the guidance of Pete Kraft & Haoyu Zhang. Available at: medrxiv.org/content/10.110…