Saketh Kapoor, PhD
@sakethkapoor
Associate Research Scientist & Lab Manager @yalemed | Spatial Proteomics Specialist | Cancer Proteomics | Single Cell Proteomics | Clearomics | timsTOF
ID: 114733281
16-02-2010 12:42:22
207 Tweet
273 Followers
279 Following

A dream of our lab has been to image the full central dogma from a single endogenous gene, all live and with single molecule resolution. After many years we are happy to unveil a beautiful cell line that makes it possible. Check out our preprint (biorxiv.org/content/10.110…)! (1/n)


Yet another must read from YingZhu (nice to see Fengchao Yu and Alexey Nesvizhskii on there too!). Impressive depth and throughput for 50µm spatial resolution. Proteome-scale tissue mapping using mass spectrometry based on label-free and multiplexed workflows biorxiv.org/content/10.110…


Want to learn how to optimize your #DDA-PASEF acquisition for #immunopeptidomics? Use Thunder-DDA-PASEF! See our paper by DavidGZ from HI-TRON Mainz: rdcu.be/dA7di Thanks to a wonderful collaboration with CompOmics and Christine CARAPITO!

Happy to contribute to this study. Group-specific FDR estimation is one of the strategies to control the error rates in proteogenomics searches for noncanonical peptides. There is tutorial on how to annotate the database and perform group FDR in #FragPipe: fragpipe.nesvilab.org/docs/tutorial_…







So excited to host Prof. Lingjun Li Lingjun Li Research Group from UW–Madison, an esteemed leader in mass spectrometry, at Yale Pharmacology!! Join us tomorrow, June 27th, at 12:30 PM in Brady Auditorium for her talk on "Advancing Biomedical Research via Innovation in MS-based Approaches." Yale School of Medicine



A protocol paper describing how to acquire and analyze multiplex DIA (mDIA) special proteomics data, with a tutorial on building a spectral library from ddaPASEF using #FragPipe. And now with diaTracer in FragPipe22, you can build a hybrid DDA+DIA library! link.springer.com/protocol/10.10…

Now online - check out this video from Karin Strube where Juliane Walz introduces our new phase I #ClinicalTrial for an #immunopeptidomics-based #peptide #vaccine for #patients with #AML. youtube.com/watch?v=tFuY4X…



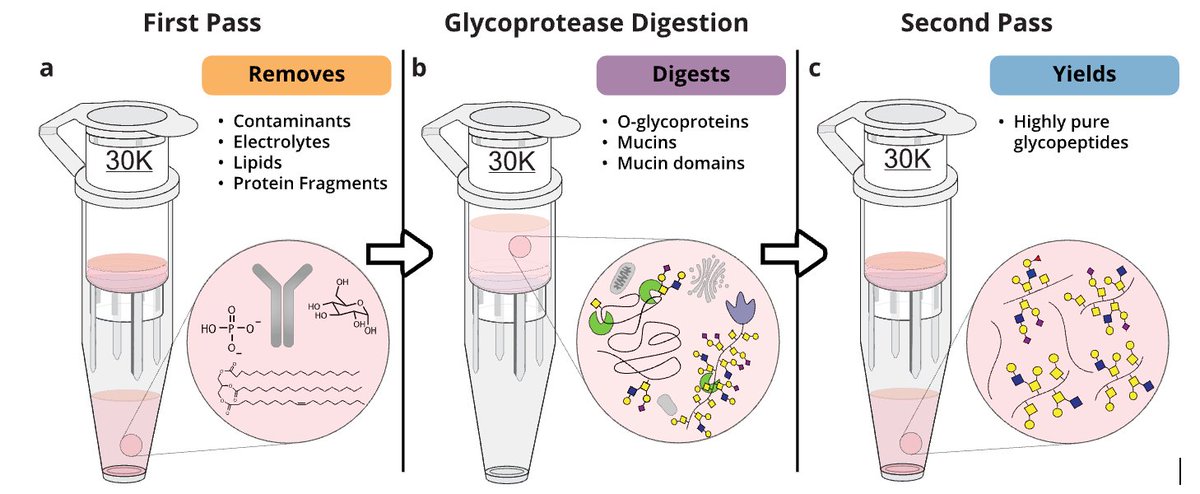
🚨New preprint alert!! Introducing "GlycoFASP" - a super easy method to isolate pure O-glycopeptides from complex samples. Pioneered by Shane and Keira E Mahoney, they show that you can simply add sample + glycoprotease to a MWCO filter -> O-glycopeptides 10.26434/chemrxiv-2025-bxv0k