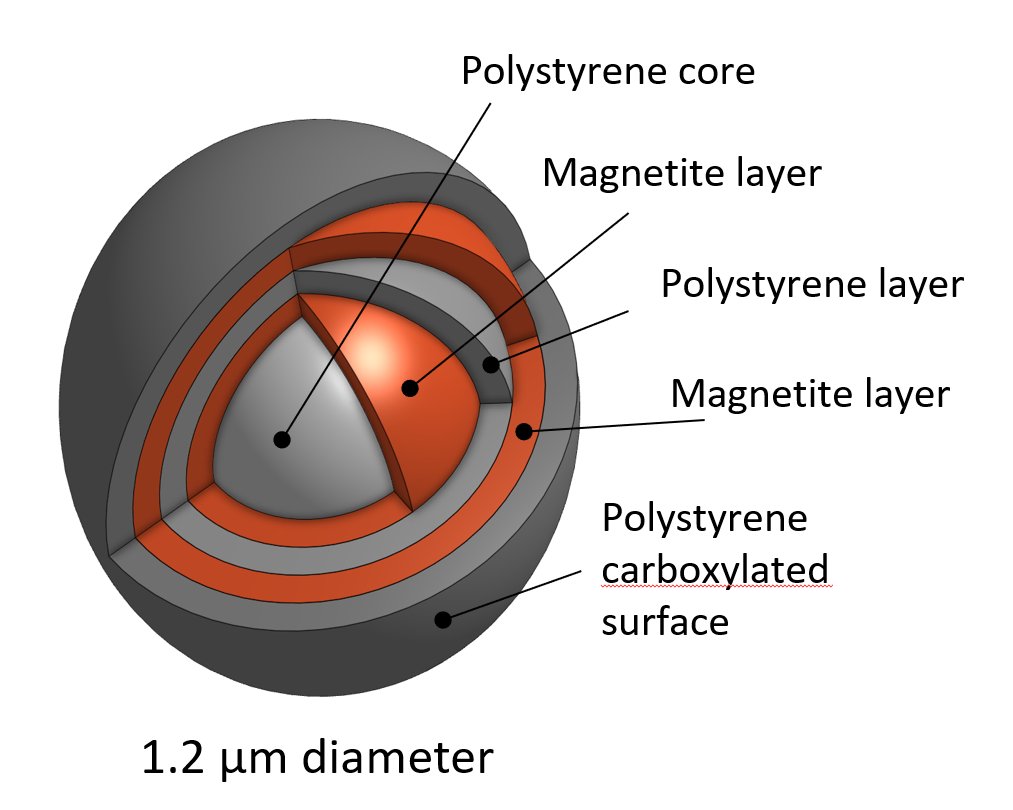
Richard Stöckl
@richard_stoeckl
PhD Student at the German Archaea Centre @uni_regensburg. #Microbiology #Bioinformatics. @[email protected]
@richard-stoeckl.bsky.social
ID: 1107942535719276545
https://richardstoeckl.gitlab.io/ 19-03-2019 09:50:59
1,1K Tweet
181 Followers
370 Following



Published in Microbiology Society tinyurl.com/4248jyuv highlighting: - Contribution of #prophage dynamics in the #pangenome evolution - Pangenome data structure to trace orthologous prophages - Generalist & specialist prophages in the Pectobacterium genus

9th de.NBI / ELIXIR-DE "Microbial Genomics training course" now open for registration! 3 sessions full of microbial bioinformatics at JLU Giessen: I: QC & QA, assembly II: regional & functional annotation III: comparative genomics Info & registration: denbi.de/training-cours…

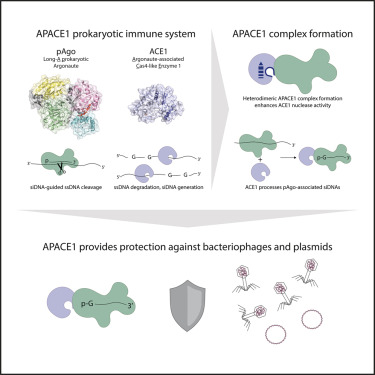
Out now in Molecular Cell: Cyanobacterial Argonautes and Cas4 family nucleases cooperate to interfere with invading DNA cell.com/molecular-cell… Most long-A pAgos interfere with invading DNA solo. Why then are cyanobacterial pAgos co-encoded with a Cas4-like protein? A 🧵

Hunting extreme microbes that redefine the limits of life book by Karen Lloyd USC Earth Science USC Marine & Environmental Biology nature.com/articles/d4158…



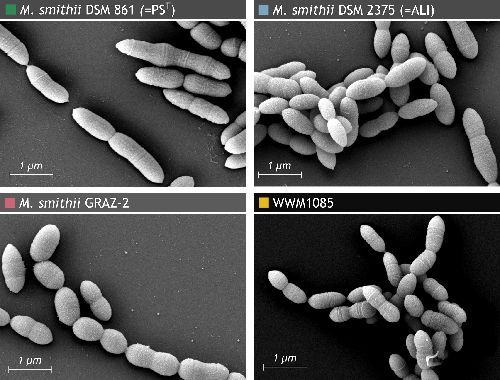
Functional prediction of proteins from the human gut archaeome #microbiology #microbiota #archaea ISME Communications ISME - International Society for Microbial Ecology academic.oup.com/ismecommun/art…







McCullough Foundation No, it did not. At best, it found that people with cancer or other diseases have different expression profiles. But it absolutely does not show any relationship with mRNA vaccines. It is a severely flawed study. Read my review here: scienceintegritydigest.com/2025/07/25/pre…