
Qin Yu
@qyucs
PhD student | bioimage analysis
ID: 1719643471
01-09-2013 16:55:05
42 Tweet
81 Followers
397 Following


Just in case you hadn’t yet noticed our supercool new toolbox for MorphoGraphX. Provides organ-centric spatial context to cellular features in 3D digital plant organs. tinyurl.com/2n6rb6ds Athul Vijayan Soeren Strauss Tejasvinee Mody Karen Lee Richard Smith Lab 🇺🇦 Plant Physiology


We have updated the manuscript to the version now accepted at CVPR (woohoo!!!). Come check it out for sparse label training in LM, EM and natural images, directly or in a domain adaptation setting: arxiv.org/abs/2103.14572. Spearheaded by Adrian Wolny , with Qin Yu and Constantin Pape.



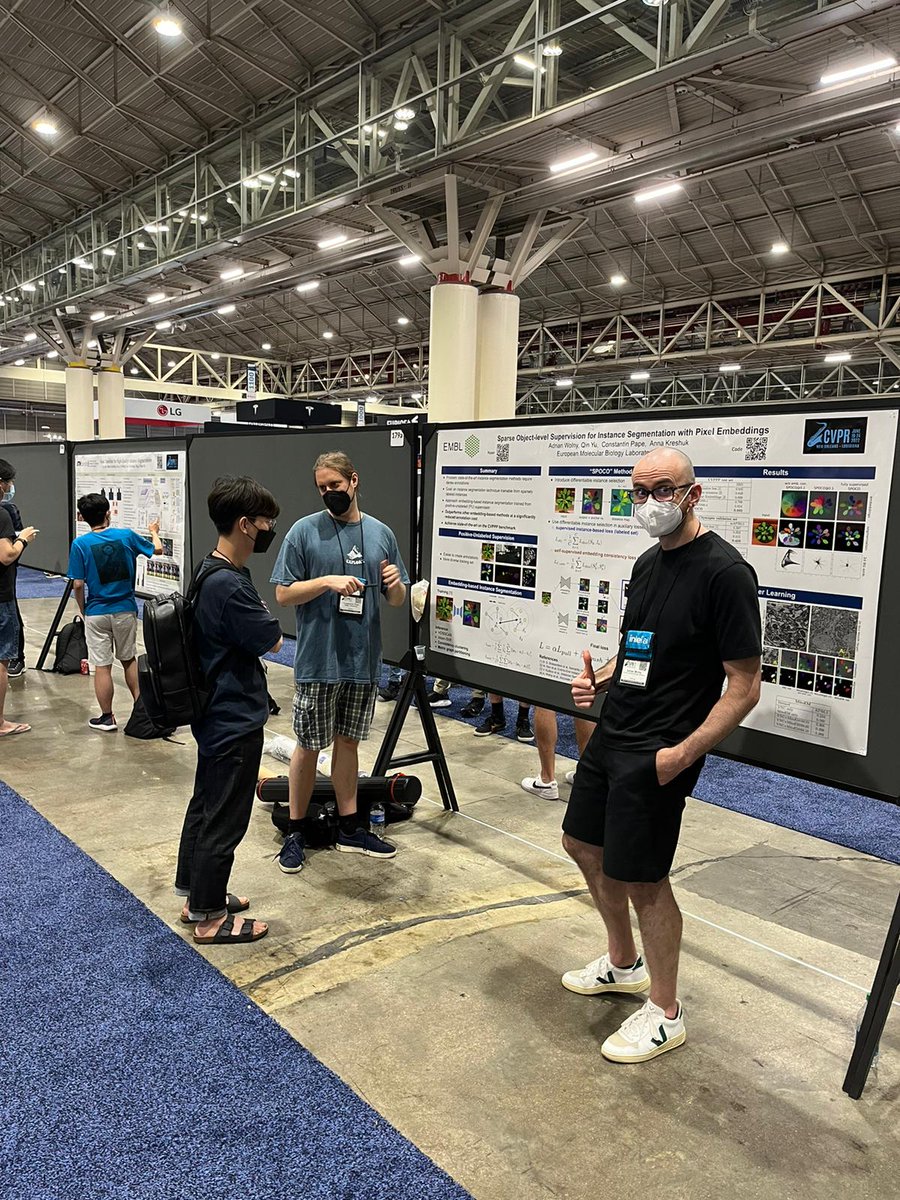
Good vibes and great discussions during the #CVPR2022 poster session. Together with Constantin Pape we presented SPOCO: our weakly supervised instance segmentation method




Finally!! My graduation ceremony at Imperial College London delayed by Covid-19 #OurImperial ✈️🇬🇧




3D nuclei segmentation using GoNuclear. Kay Schneitz Tejasvinee Mody Qin Yu Soeren Strauss Adrian Wolny Lorenzo Cerrone Fred Hamprecht Richard Smith Lab 🇺🇦 #Anna_Kreshuk github.com/kreshuklab/go-… biorxiv.org/content/10.110…

Just out in Development. With our Go-Nuclear models, it is now possible to reliably 3D segment nuclei from 3D digital images. bit.ly/4dcYBz0 bit.ly/3zLS3ZO Athul Vijayan, Tejasvinee Mody, Qin Yu, Soeren Strauss, Adrian Wolny, Lorenzo Cerrone, Richard Smith Lab 🇺🇦

A deep learning-based toolkit for 3D nuclei segmentation and quantitative analysis in cellular and tissue context Read this Techniques and Resources Article by Athul Vijayan, Tejasvinee Mody, Qin Yu, Kay Schneitz Kay Schneitz and colleagues: journals.biologists.com/dev/article/15…




