
Jan-Philipp Quast
@quastjp
PhD student in the @Picotti_Lab at ETH Zurich. Author and maintainer of protti. jpquast.github.io/protti/
ID: 998190468004446208
20-05-2018 13:15:27
87 Tweet
184 Followers
203 Following
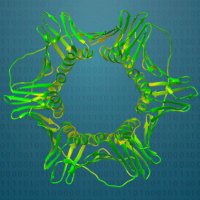

Very proud to announce that my first publication was just published in Bioinformatics Advances! Please check out the paper doi.org/10.1093/bioadv… Also check out our R package 📦: jpquast.github.io/protti/




So great to see this story out! Really proud to have been able to contribute to this amazing project during my time in the lab! Congrats to Max Valenstein, MD, PhD and @kbrogala for shedding light on the structure and function of GATOR2 a truly fascinating protein complex!

Super excited to share this collaborative work nature that focuses on the roles of Sestrin-mTORC1 in organismal dietary leucine response. So much fun working with Patrick Jouandin from Perrimon Lab. Thank all co-authors for their very valuable contributions.nature.com/articles/s4158…



Check out our new protti release (0.5.0) here ➡️ jpquast.github.io/protti/ Jan-Philipp Quast was a busy 🐝, fixing some bugs and implementing the predict_alphafold_domain() function based on Tristan Croll‘s Python Code.


Big thanks to Brady Johnston for a great first #Blender workshop day! I learned a lot and could even already render a very simple but pretty animation! 🤩

Jan-Philipp Quast I've tried it in my Shiny app for qPCR analysis and seems impressive. Thank you. BTW, it fully supports facet_wrap().



Recent protti updates 👩🏻💻 My former colleagues of the Picotti Lab Jan-Philipp Quast & Elena Krismer have been busy fixing bugs & implementing new features in our R package. Lots of improvements & additional arguments for more flexibility. Check them out here ⬇️ jpquast.github.io/protti/news/in…


