Pranam Chatterjee
@pranamanam
Designing biologics to program biology! 💻🧫 Assistant Professor at @DukeU | Co-Founder @GametoGen and @UbiquiTxINC | Formerly @MIT '16 '18 '20 and @harvardmed
ID: 3404225795
http://programmable.bio 05-08-2015 14:08:57
572 Tweet
3,3K Followers
326 Following

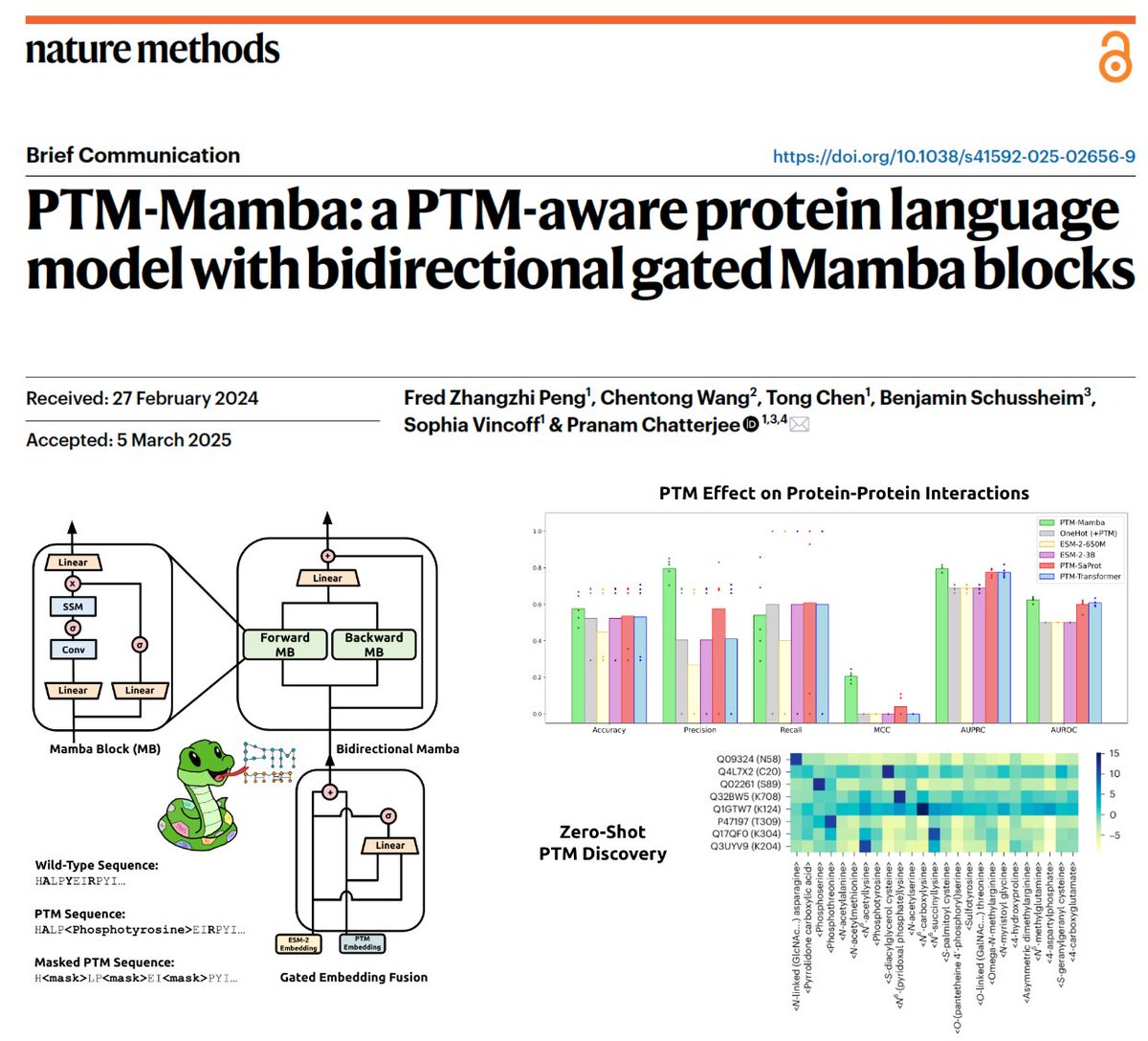
PTM-Mamba is out in Nature Methods!! 🥳 PTM-Mamba is the first/only pLM to encode both WT and post-translationally modified residues as tokens! And we do this without losing any base protein information from ESM-2! 🤝 📜: nature.com/articles/s4159… 💻: github.com/programmablebi… We

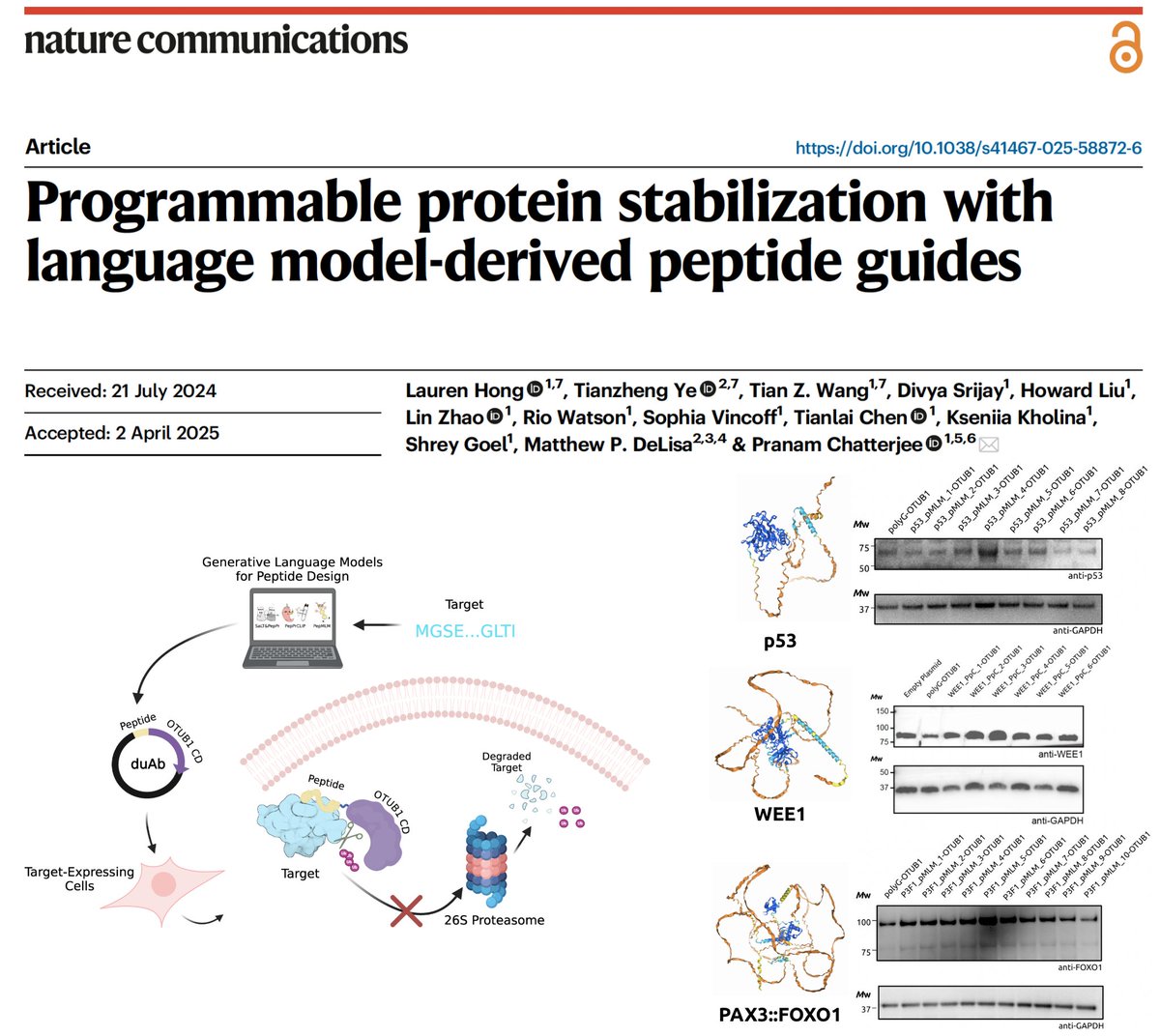
Our pLM-designed peptides, when attached to E3 ligases (aka uAbs), enable us to programmably degrade disease targets (think CRISPRi, but for proteins). But what if we want to STABILIZE useful proteins? In Nature Communications, we introduce duAbs! 🌟 📜: nature.com/articles/s4146… 🧬:

Super excited to be at #ICLR2025 in Singapore! 🇸🇬 My students and I are presenting 20+ accepted works across the workshops/main, including at ICLR Nucleic Acids Workshop Learning Meaningful Representations of Life #DeLTa #FPI! We're especially excited to co-host the GEMBio Workshop tomorrow!! 💻🧬🧫 Come say hi!! 👋



This was one of my favorite experimental papers from our GEMBio Workshop at ICLR!! 🇸🇬 A super crafty way of generating longer DNA libraries via Golden Gate -- we'll definitely be using it for our proteome editing screens!! 🧬 Really great work Shaozhong Zou!! 👏


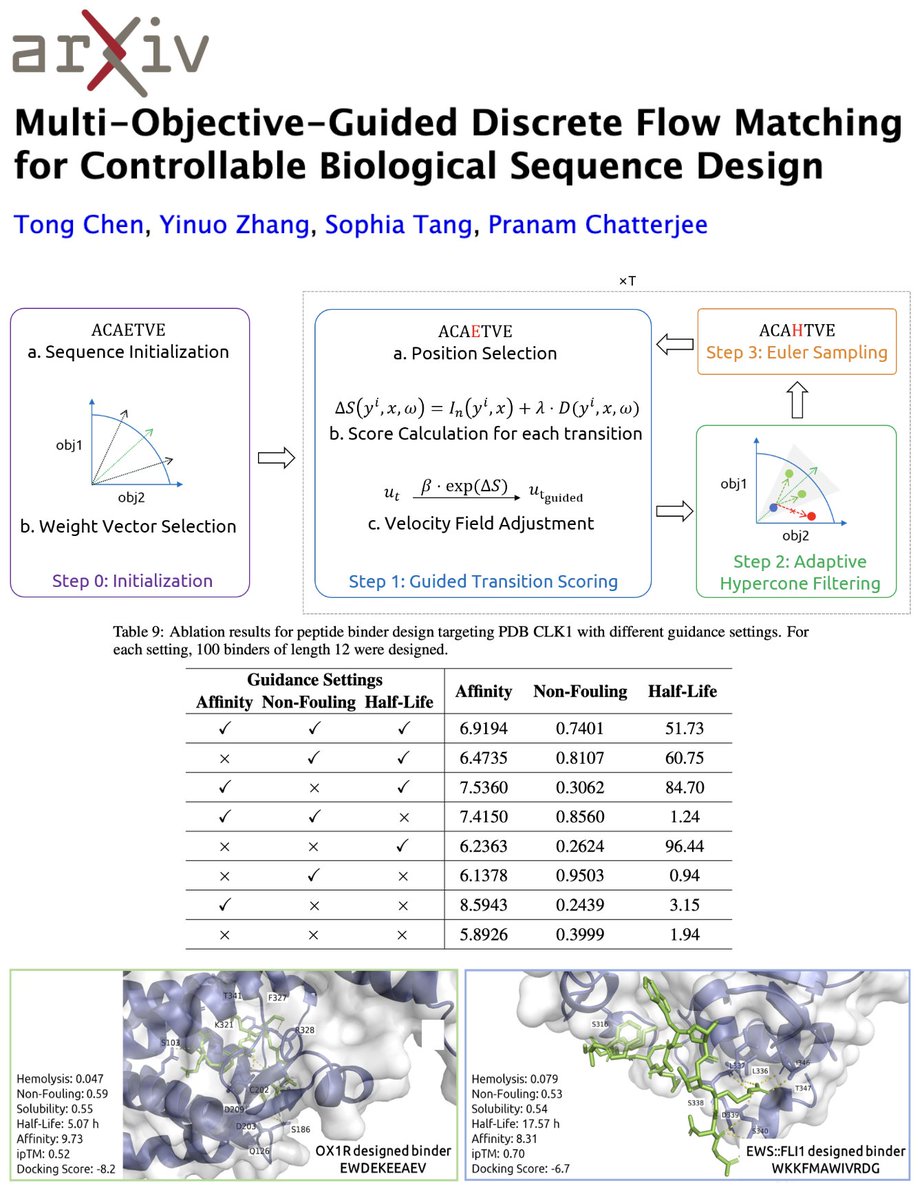
As you know, we’ve been deep in the discrete diffusion trenches for sequence design for quite some time now (both theoretically and for biology!)—PepTune for multi-objective discrete diffusion for therapeutic peptide design from Sophia Tang, P2 for path planning and improved

I absolutely love this paper!! 🌟 Beautiful work by Jason Yang Yisong Yue and team to show that discrete diffusion models can be effectively guided by fitness data from low-throughput wet-lab assays! 🧪 I'm telling you guys: guided sequence generation is where it's at! 🤙


Most biological processes, like stem cell differentiation, branch into multiple fates, but current trajectory inference methods only predict single paths.🌳To fix this, we introduce BranchSBM 🌿, from our unstoppable Sophia Tang ! 🌟 📜: arxiv.org/abs/2506.09007 💻: