OmniPath
@omnipathdb
OmniPath: molecular biology knowledge from 100+ databases
ID: 923168730502369280
http://www.omnipathdb.org 25-10-2017 12:45:51
68 Tweet
407 Followers
30 Following

Check out my first first-author work with Manish Butte! We used OmniPath, a protein interactions meta-database, as well as RNAseq data from Human Protein Atlas to expand the potential genes of inborn errors of immunity (IEIs). Hoping immunology clinics worldwide will find it useful!



Human Cell Map from gingraslab is available in OmniPath! ⭐️ Subcellular localization of 6,000+ proteins from a BioID screen in HEK cells: cell-map.org, nature.com/articles/s4158…


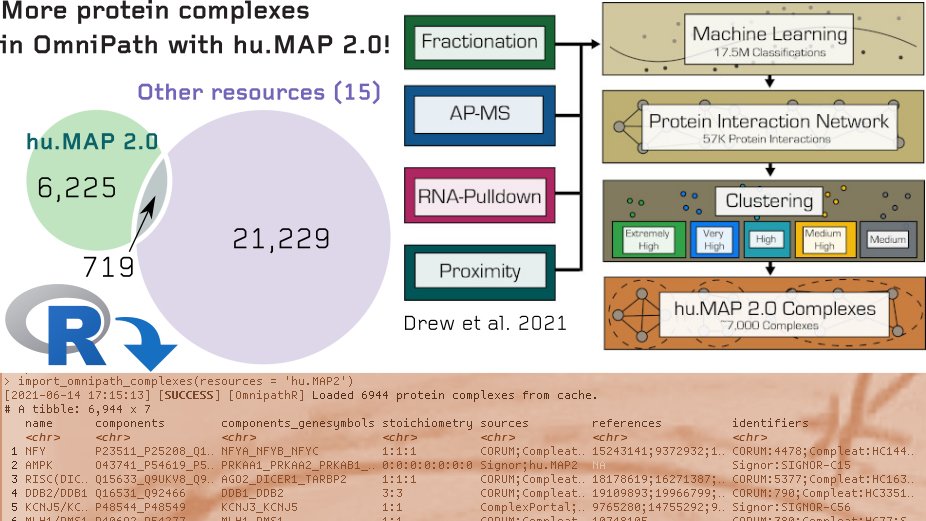
6,225 new protein complexes in OmniPath thanks to hu.MAP 2.0 from Kevin Drew John Wallingford & Edward Marcotte 🖥️ omnipathdb.org/complexes?reso…, 🏡 humap2.proteincomplexes.org, 📰 embopress.org/doi/full/10.15…

11 #PPI scores based on structure, sequence and function from PrePPI of Honig Lab Columbia University 🌴honiglab.c2b2.columbia.edu/PrePPI 🌴 Now easy access with our R package: static.omnipathdb.org/omnipathr_manu… 🌴 github.com/saezlab/Omnipa…


We just talked about OmnipathR at BioC2021 Bioconductor, many thanks to all the participants and the organizers! 🌞 Slides: static.omnipathdb.org/bioc-omnipath_… Vignette (and further materials): saezlab.github.io/OmnipathR/arti… Other Saez-Rodriguez Group tools: saezlab.org/#tools

Check out #COVID19 Disease Map Consortia COVID-19 Pathways knowledge repository embopress.org/doi/full/10.15…, great community effort where we contributed OmniPath (pathway knowledge), DOROTHEA saezlab.github.io/dorothea (TF targets) & CARNIVAL saezlab.github.io/CARNIVAL (network modeling)

Our paper is among the 8 most read in Mol Syst Biol embopress.org/doi/full/10.15…, and got already 27 citations! 🎉 In Saez-Rodriguez Group & KorcsmarosLab it's a great pleasure for us to see you using our work. Meanwhile we are preparing big new developments, stay tuned 😉


The great spatial omics platform from Fabian Theis group is out in Nature Methods 🥳 Squidpy uses OmniPath data for ligand-receptor interactions 🤩 Special thanks to Michal Klein for implementing the OmniPath Python client✌️



We are looking for PostDosc/ PhDs in Medical Bioinformatics & Machine Learning Universitätsklinikum Heidelberg Uni Heidelberg #multiomics #singlecell consider applying and pls RT 🙏 jobrxiv.org/job/heidelberg…

Check MISTy from Jovan Tanevski et al Saez-Rodriguez Group to analyze spatial omics - they used OmniPath 's annotated prior knowledge to link intracellular pathway activities & the expression of ligands in space 👇