
MolecularGenetics@ErasmusMC
@moleculargenet3
ID: 1367449313727627268
04-03-2021 12:19:13
172 Tweet
178 Followers
111 Following


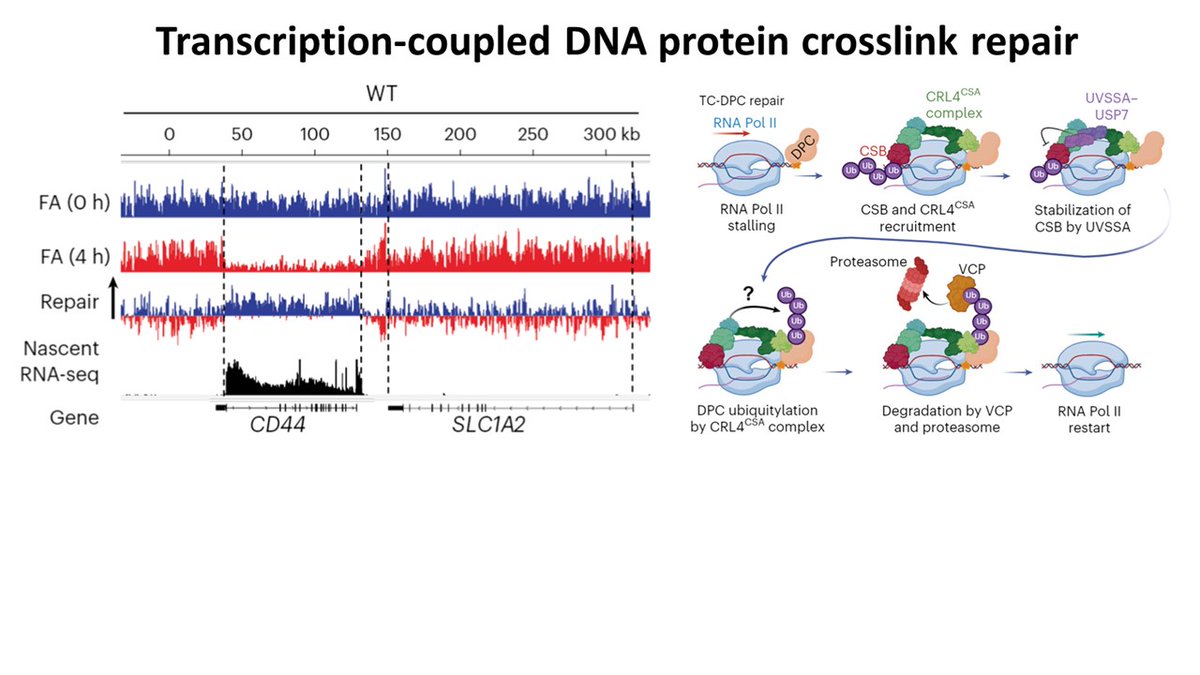
Thrilled to announce our latest research in Nature Cell Biology! We uncovered the crucial role of CSA and CSB proteins in repairing DNA-protein crosslinks (DPCs) in a novel transcription-coupled DPC-repair pathway. Erasmus MC MolecularGenetics@ErasmusMC Oncode Institute nature.com/articles/s4155…


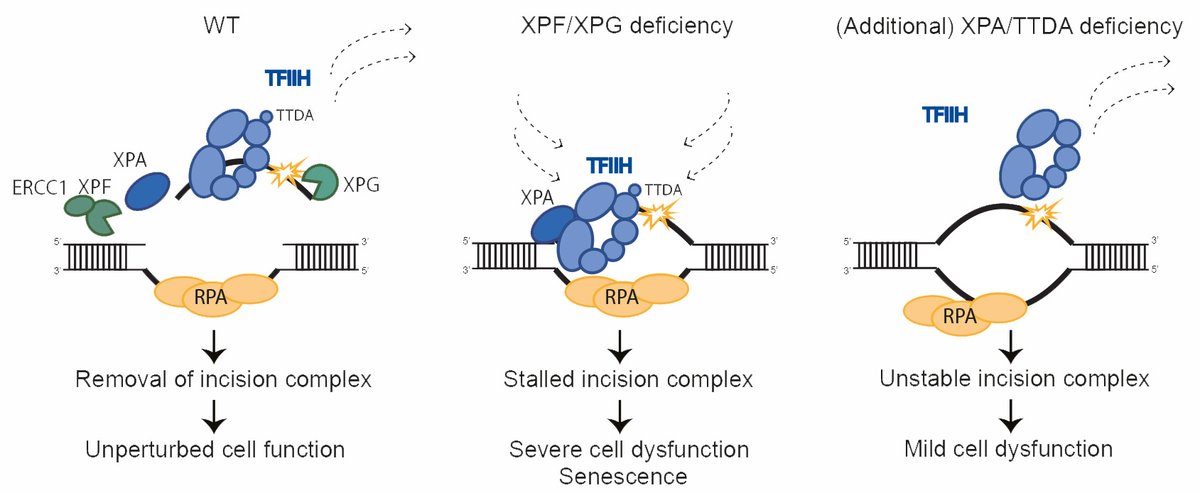
Happy to share our Nature Communications paper showing that prolonged TFIIH binding to nonexcised DNA damage causes severe neuronal and developmental failure. This can be prevented by depletion of XPA or TTDA, which stabilize TFIIH-DNA binding. nature.com/articles/s4146…


Happy to share our newest preprint on the function of the new TC-NER factor STK19. Great work of Anisha and Shun and a very nice collaboration with the Sixma lab @TitiaSixma. Erasmus MC MolecularGenetics@ErasmusMC Oncode Institute biorxiv.org/content/10.110…


Nitika Taneja we are looking forward to your presentation at #BGFP25 in Portugal next July🌞! Registration is OPEN so please RT to let your followers know! For more info -🔗bit.ly/3AnsVZF







Congratulations Sounak Sahu, Mélissa Galloux, members of our BRCA Variant Analysis Unit National Cancer Institute for this outstanding work. Sincere thanks to our collaborators Drs. Chari, Papaleo, Michailidou and their team members.








