
Lisa Baumgartner
@lisababalisa
Fascinated by how specificity is achieved at chromatin!
PhD with @juliusbrennecke @IMBA_Vienna #smallRNAs 🪰🦏🔬
Postdoc @SchubelerLab @FMIscience #TFs 🐁🧫🧬
ID: 900281544945917952
23-08-2017 09:00:21
208 Tweet
321 Followers
389 Following


The @juliusbrennecke group shares an update on last year's Kipferl discovery by Lisa Baumgartner.




Yay! Super happy to see my paper out in EMBO Reports. Thanks to all the people involved in the work, the reviewers and the editor. For a summary, check out the interview below 👇 or this threat 🧵: x.com/llorenzoorts/s…


Peters Lab: Excited to share our latest preprint lead by Greg Fanourgakis (Grigo Fanou). Key findings are that DNA methylation modulates nucleosome retention in sperm and H3K4 methylation deposition in early mouse embryos. biorxiv.org/content/10.110…


We're looking for a Managing Director of our scientific core facilities - details: viennabiocenter.org/fileadmin/user…. Come work with us - 6 basic research institutes & 40 biotech companies, >2000 scientists from 85 nations - at the vibrant Vienna BioCenter in Vienna, Austria. Please RT.

I’m beyond excited to present my postdoc work with Plaschka Lab and @JuliusBrennecke at IMP IMBA imbavienna.bsky.social . We uncovered a mechanistic framework for a general and conserved mRNA nuclear export pathway. A thread: 1/x


Congratulations to Julius Brennecke for securing one of this year’s European Research Council (ERC) Advanced Grants! Julius will study the arms race between transposons and host genomes. #ERCAdG More here: oeaw.ac.at/imba/research-…

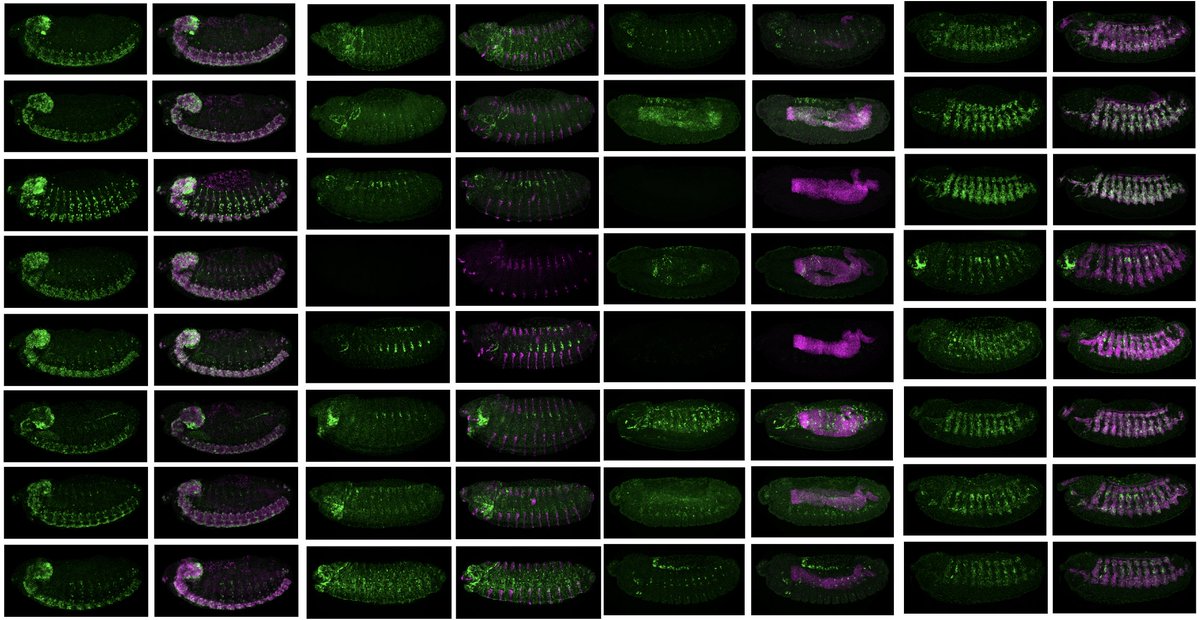
Excited to share our latest preprint. Lead by Sevi (Sevi Durdu) we asked how transcription factors (TFs) with short and abundant motifs drive specific cell fates. Our test cases: Two bHLH factors that bind to seemingly similar E-boxes. 1/7 biorxiv.org/content/10.110…


It is challenging to study the ability of transcription factors to engage their motifs in the cell. We tried to address this in our new study together with Marco (Marco Pregnolato), Schübeler Lab, and FMI science: cell.com/molecular-cell…

To pioneer or not to pioneer...! Or is it a bit more complicated after all? Check out the latest from the Schübeler Lab, a huge effort by Marco Pregnolato and Ralph Grand 🎉 (featuring the first small snippets of my PD work 😊)

Cells turn genes on and off with the help of transcription factors, but binding to DNA is complex. Marco Pregnolato, Ralph Grand and others in the Schübeler Lab found that these factors act more variably than expected, sometimes displacing nucleosomes only in certain regions 🧬
![Ibrahim I. Taskiran (@ibrahimihsan) on Twitter photo Exciting news! Our latest research on "Cell type directed design of synthetic enhancers" has been published in <a href="/Nature/">nature</a>. We've harnessed deep learning to design enhancers with cell-type specificity. Find our paper here: nature.com/articles/s4158…. Highlights below: [1/n] Exciting news! Our latest research on "Cell type directed design of synthetic enhancers" has been published in <a href="/Nature/">nature</a>. We've harnessed deep learning to design enhancers with cell-type specificity. Find our paper here: nature.com/articles/s4158…. Highlights below: [1/n]](https://pbs.twimg.com/media/GBKA6x7XQAAn_7m.jpg)