Hofmann_Lab
@labhofmann
Research group at the University of Cologne
ID: 1282701583948681226
13-07-2020 15:41:26
54 Tweet
165 Followers
110 Following

In our new preprint, we show that a Chlamydia-like bacterium (Simkania) has a deubiquitinase repertoire similar to that of Legionella, but using very different DUB classes for the same linkage specificities (e.g. M1, K6). #ubiquitin Universität zu Köln researchsquare.com/article/rs-264…





Out now in NatComms: A Chlamydia-like bacterium with a deubiquitinase repertoire similar to Legionella, but using new DUB classes for the same linkage specificities (e.g. M1, K6). #ubiquitin Universität zu Köln Cologne Crystallisation facility doi.org/10.1038/s41467…



By the way: this project was the brainchild of excellent postdoc Thomas Hermanns, who was also corresponding author! We also acknowlegde support from Cologne Crystallisation facility and Universität zu Köln






New Lammers Lab paper out now in Nature Communications!🥳 We reveal high functional diversity within bacterial deac(et)ylases by systematic functional/structural characterization. This is Leonies PhD project and I’m grateful to have contributed to this great work. nature.com/articles/s4146…

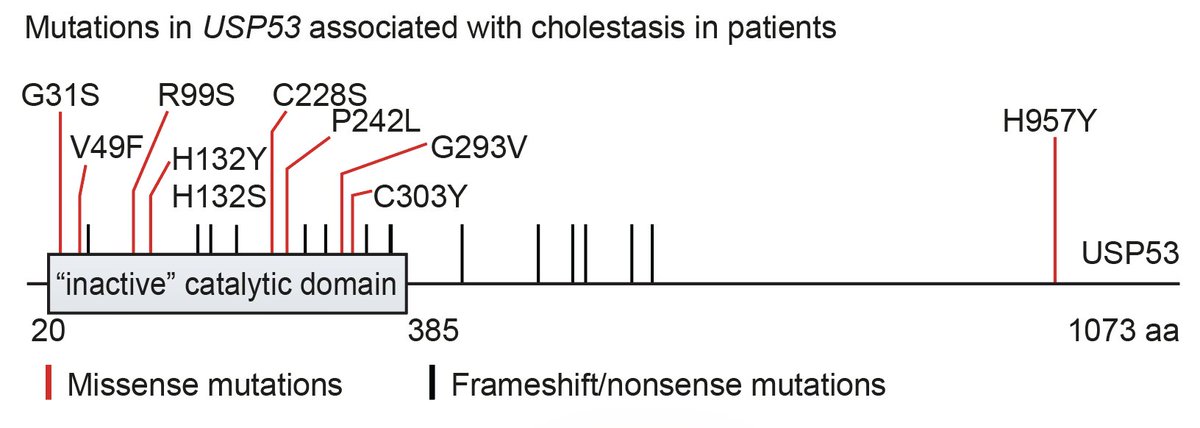
Very excited to share that our manuscript on the "Discovery and mechanism of K63-linkage-directed deubiquitinase activity in USP53" was published today at Nature Chemical Biology Nature Chemical Biology. Congratulations Kim, Kai, and all authors! nature.com/articles/s4158…