Laboratory of biomolecular modelling and design
@kullbmd
Arnout Voet, Computational protein and drug design, structural biology, biomolecular modelling, protein crystallography and biophysics.
ID: 1272077612303728640
14-06-2020 08:05:43
309 Tweet
738 Followers
417 Following

Warm congratulations to PhD student Ahmed Shemy Laboratory of biomolecular modelling and design on winning the IUCr Journals Poster Prize at the 10th Diamond Light Source ccp4_mx Data Collection and Structure Solution Workshop, shown receiving his certificate and prize from lecturer and Structural Biology Editor Elspeth Garman


DCU School of Chemical Sciences Dublin City University Hi all. I hope you can please #RT this? Jan Jensen (janhjensen.bsky.social) Peter Kenny 🇹🇹🇬🇧🇧🇷🇺🇦🌻🇪🇺🇲🇦 Chris Swain Noel O'Boyle (@[email protected]) Matthieu Schapira @EstebanezLab Laboratory of biomolecular modelling and design Cresset CCG_MOE OpenEye, Cadence Molecular Sciences MGMS Thanks for passing this along.

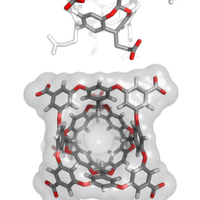


In our latest paper we engineered a nuclear receptor ligand specificity to create a new synbio tool. Congrats to all authors. Thanks for the great collaboration with Molendo Kevin Verstrepen lab doi.org/10.1002/pro.49… Fac Wetenschappen KULeuvenChemie


Sometimes #AFM gives you this joy! micrometer wide Protein-based 2D layers made from a Computational Designer protein. Excited to see where this protein will lead us next! #BioAFM #AtomicForceMicroscopy #CPD #proteindesign #Nanobiotechnology Laboratory of biomolecular modelling and design KU Leuven #biochemistry
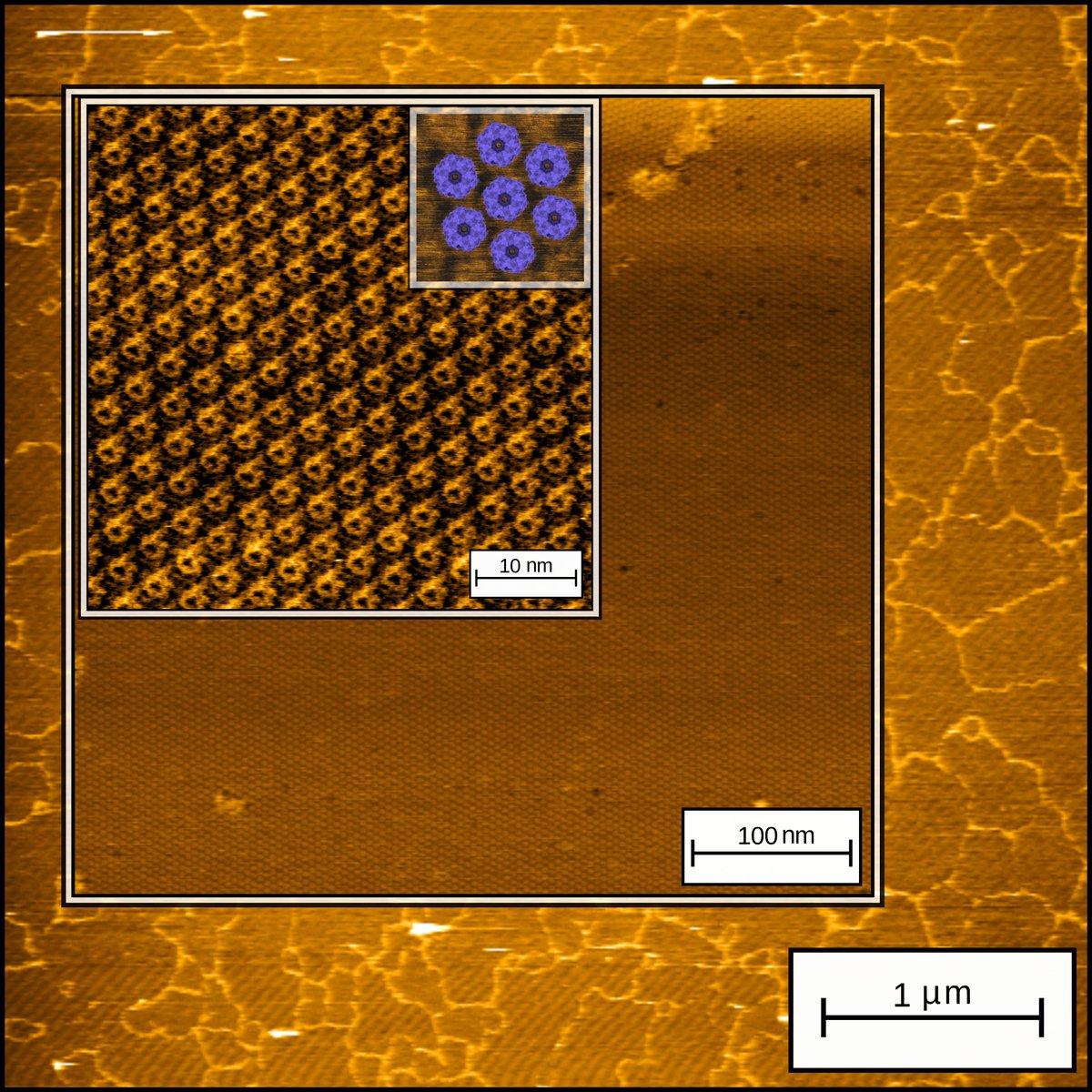

protein layers as far as the #AFN eye can see (+5um) : Great work of one of our PhD students working on development of functional #biomaterials through computational protein design and #AFM. Stay tuned for more Fac Wetenschappen KULeuvenChemie

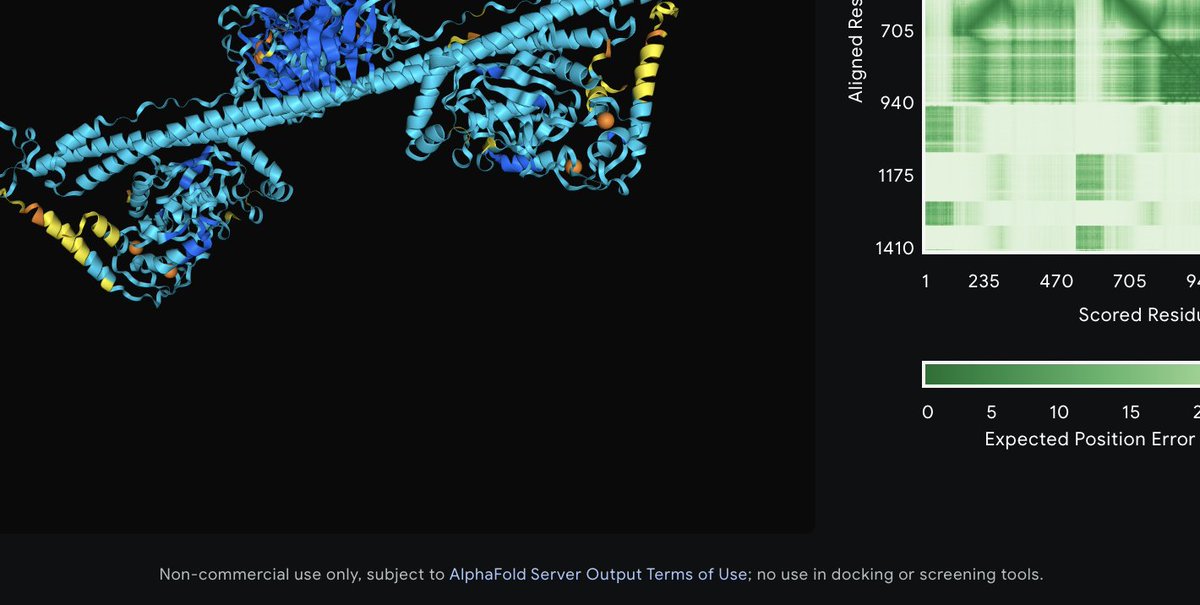
I just noticed the little warning underneath the results of an AF3 job. OK, so Google DeepMind will never let us use their ligand binding diffusion model. But you can't even use their protein models for ligand docking. That's ridiculous.






Prof Xin Shi on his #ERCStG: "I'm very grateful that there are funding opportunities in the EU for curiosity-driven basic science. It’s a recognition of the importance to boldly try things that no one has before." European Research Council (ERC) Fac Wetenschappen KU Leuven set.kuleuven.be/en/news/interv…