Jue Wang
@jueseph
Research scientist @googledeepmind. Formerly @UWproteindesign @ginkgo @HMS_SysBio.
ID: 25824880
http://juewang.strikingly.com 22-03-2009 14:43:16
2,2K Tweet
2,2K Followers
770 Following

With David and the Baker Lab in the spotlight today, I wanted to share some insights into the Institute for Protein Design and how it operates, a glimpse behind the curtain. I had planned to write this post-graduation, but now seems as good a time as any. (Got twitter blue free trial so this


Now online: We created de novo insulin receptor agonists using computational protein design! Choi Lab Xiaochen Bai Preetham Venkatesh Institute for Protein Design #BakerLab biorxiv.org/content/10.110…


Finally, out in Science Magazine today, our work on de novo protein design. Together with Motoyuki Hattori (服部素之) , Sergey Ovchinnikov & @hendrik_dietz we showed how we can drastically improve AF2 based hallucination, surpassing, or reaching SOTA diffusion models doi.org/10.1126/scienc…

Remember Golden Gate Claude? Etowah Adams and I have been working on applying the same mechanistic interpretability techniques to protein language models. We found lots of features and they’re... pretty weird? 🧵



In a new article for nature, journalist Sara Reardon explores “Five structural-biology questions that still challenge AI,” along with a number of experts in the field including John Ingraham, our Head of Machine Learning, Alena Khmelinskaia, David Baker (University of Washington), Jue Wang,



The #AlphaFold 3 model code and weights are now available for academic use. We Google DeepMind are excited to see how the research community continues to use AlphaFold to address open questions in biology and new lines of research. github.com/google-deepmin…



After showing that AF2 can be used to design very large proteins by performing #RSO, I am happy to share another fun project we did: the #af2cycler Sergey Ovchinnikov @hendrik_dietz biorxiv.org/content/10.110…



Deep mutational scanning of Rubisco, from Noam Prywes Savage Lab (I really liked the note in the "Behind the paper" section): nature.com/articles/d4158…




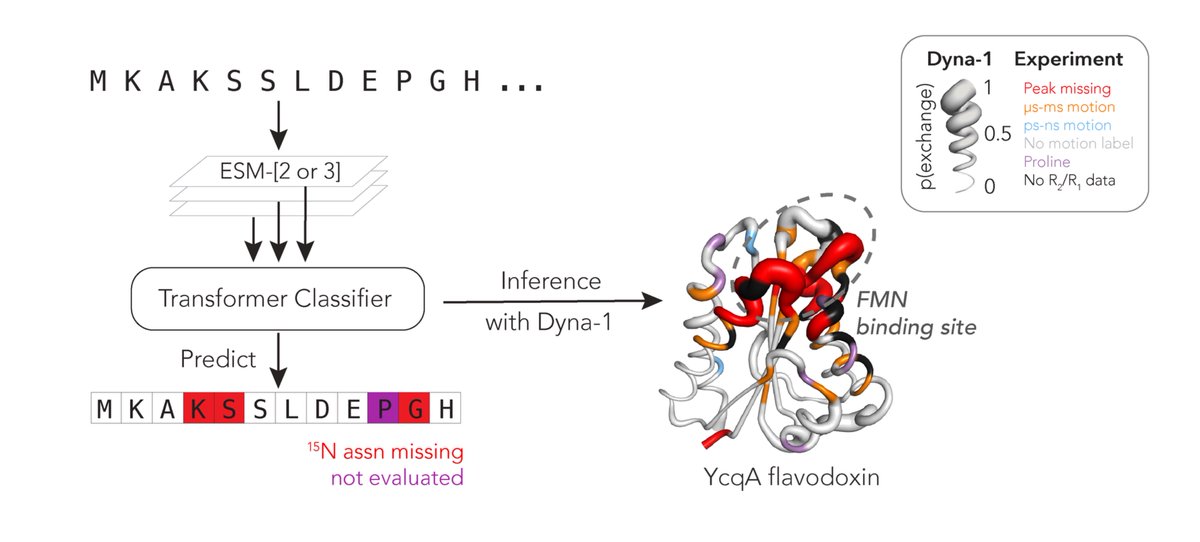
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics? Hannah Wayment-Steele and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1! 🧵