Wook 🐪
@joowook
Clinically Bioinformatically Genomically
ID: 21603637
22-02-2009 22:04:45
1,1K Tweet
453 Followers
1,1K Following

📢STP Equivalence funding available for 200 healthcare scientists 📢 Along with the Academy for Healthcare Science we are delighted to announce an exciting initiative that aims to increase the number of statutory registered healthcare scientists. ℹ️ nshcs.hee.nhs.uk/news/stp-equiv…


Huge congratulations to Prof Matt Hurles (Matt Hurles ) who has been appointed as the new Director of Wellcome Sanger Institute! 🎊 Currently Head of the Human Genetics Programme, Matt will formally take up the Directorship later this year. Find out more⤵️ bit.ly/3Xq3T26





What is an exon??? New paper w/ Dr. Edward Wallace and julie aspden in Cell Genomics : cell.com/cell-genomics/… Cue 🧵1/8





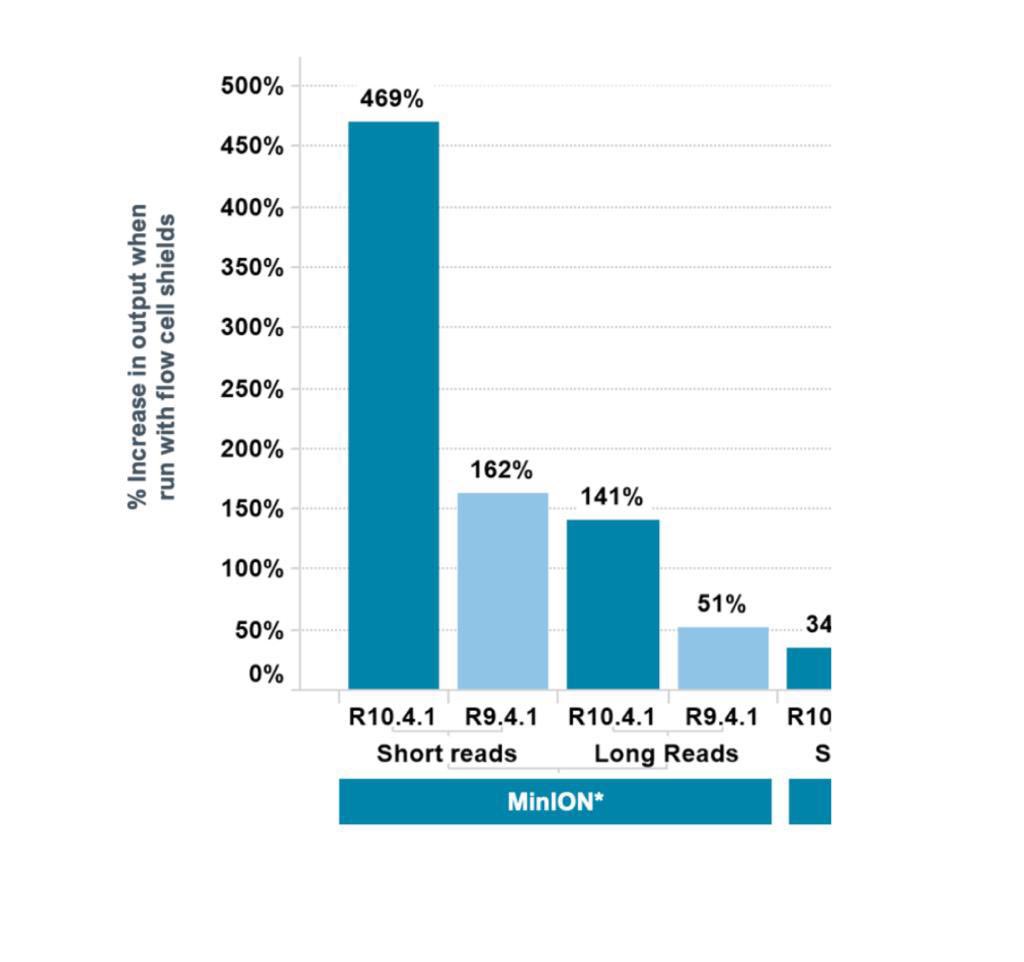
So Oxford Nanopore discovered something a few days ago - and it has to do with light. And they posted this image (sorry, too good not to share off the community). Yes, that’s 469% increased output. So, we decided to give it a shot and …