Harm-Jan
@harmjanwestra
Postdoc @ University Medical Center Groningen
ID: 39512144
12-05-2009 14:38:23
99 Tweet
214 Followers
197 Following

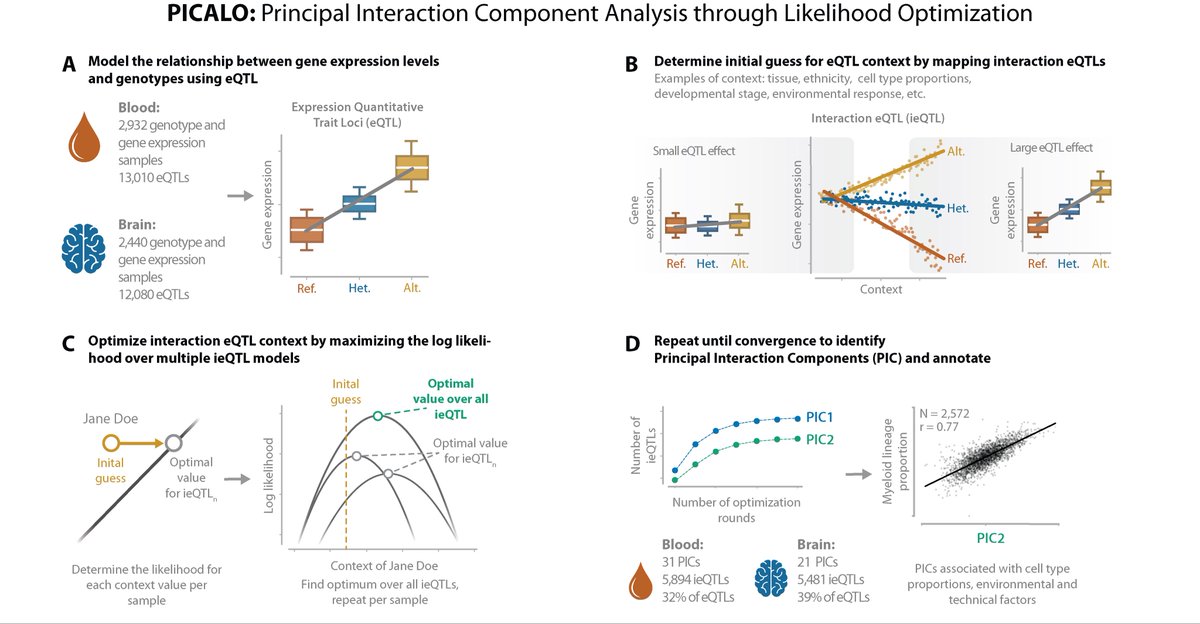
‼️ ONLINE NOW Orli Bahcall 📰 Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases 🧑🤝🧑 Heiko Runz Lude Franke Harm-Jan and colleagues ⬇️ go.nature.com/3ZosKoC






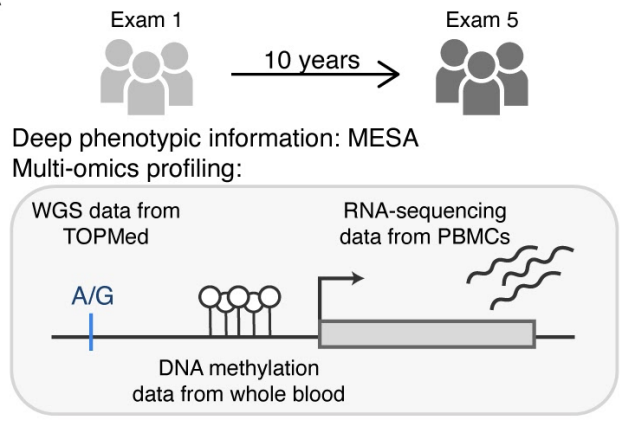
Excited to share a new preprint from my lab, led by Silva Kasela ✨, analyzing cellular and environmental modifiers of regulatory variants in the TOPMed multi-omics pilot data from the MESA cohort. 🧵biorxiv.org/content/10.110…

We are very proud that Monique van der Wijst Monique van der Wijst has been awarded a VIDI grant for her project to study gene expression variation. Monique, many congratulations! UMCG University of Groningen NWO Nieuws

I am very happy and grateful to tell you that I've been awarded a NWO Vidi grant to study the role of non-genetic molecular variation among seemingly identical immune cells for immune cell memory, and its relation to disease: shorturl.at/kAIJN UMCG Genetica Research UMCG

Very proud of my colleague Monique van der Wijst !


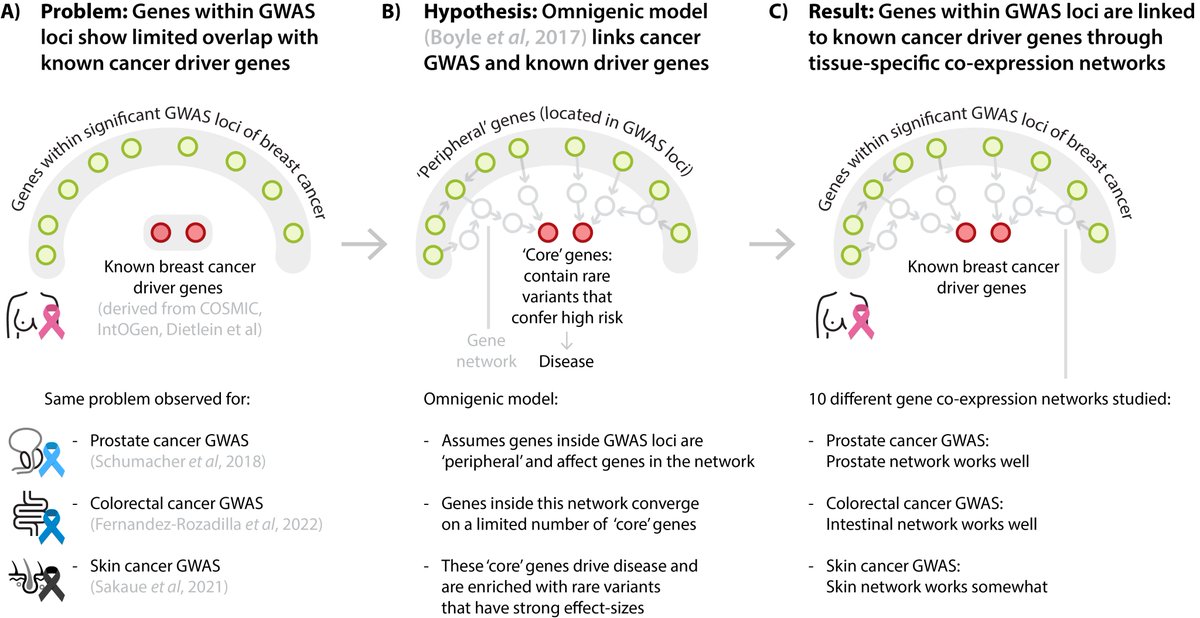
The overlap between GWAS hits in cancer and established somatic driver genes is quite low. We observe that tissue-specific co-expression networks link the positional candidate genes in cancer GWAS to known somatic driver genes. Tijs van Lieshout explains below what we found: