
gagneurlab
@gagneurlab
News from the Gagneurlab@TUM -- To understand the genetic basis of gene regulation and its implication in diseases.
ID: 832179311062945794
https://www.in.tum.de/gagneurlab 16-02-2017 10:46:23
445 Tweet
1,1K Followers
111 Following

Sequence design at the upcoming Kipoi webinar on Wed :-) Looking forward to Sager Gosai's talk!

Join us for an exciting afternoon of RNA talks (including Giulia Cantini @[email protected] from my lab) on the 10th of July at MPI for Biochemistry! We will celebrate summer with some pizza and drinks afterwards Computational Health Center MUDS




After DeepRVAT, FuncRVP is our next rare variant association testing tool! Rather than testing one gene at a time, we leverage information across genes using foundation models of gene function. All open source. Shubhankar Londhe Shubhankar Londhe led this cool work - more in his thread



Join us for the next Kipoi Seminar ft. Max Horlbeck & Ruochi Zhang (Ruochi Zhang) from Jason Buenrostro's lab at Harvard University & Broad Institute! 👉Single-cell footprinting reveals the organization of cis-regulatory elements 📆Wed Sep 4, 5:30pm CET 🧬tum-conf.zoom.us/meeting/regist…


Taking sequence-based modeling to the next level: 500 kb DNA input, single-cell resolution, joint ATAC & RNA coverage tracks. Thumbs up to Johannes Johannes Hingerl & Laura Laura Martens (@lauradmartens.bsky.social). Wonderful collab with Jason Buenrostro & Fabian Theis. Open source, of course. Enjoy!

Really pleased to see DeepRVAT published. A new method for rare variant genetics by Eva Holtkamp & Brian Clarke in collab with gagneurlab. I like to think of DeepRVAT as one network on top of another, harnessing variant annotations from bags of models- AlphaMissense and alike.

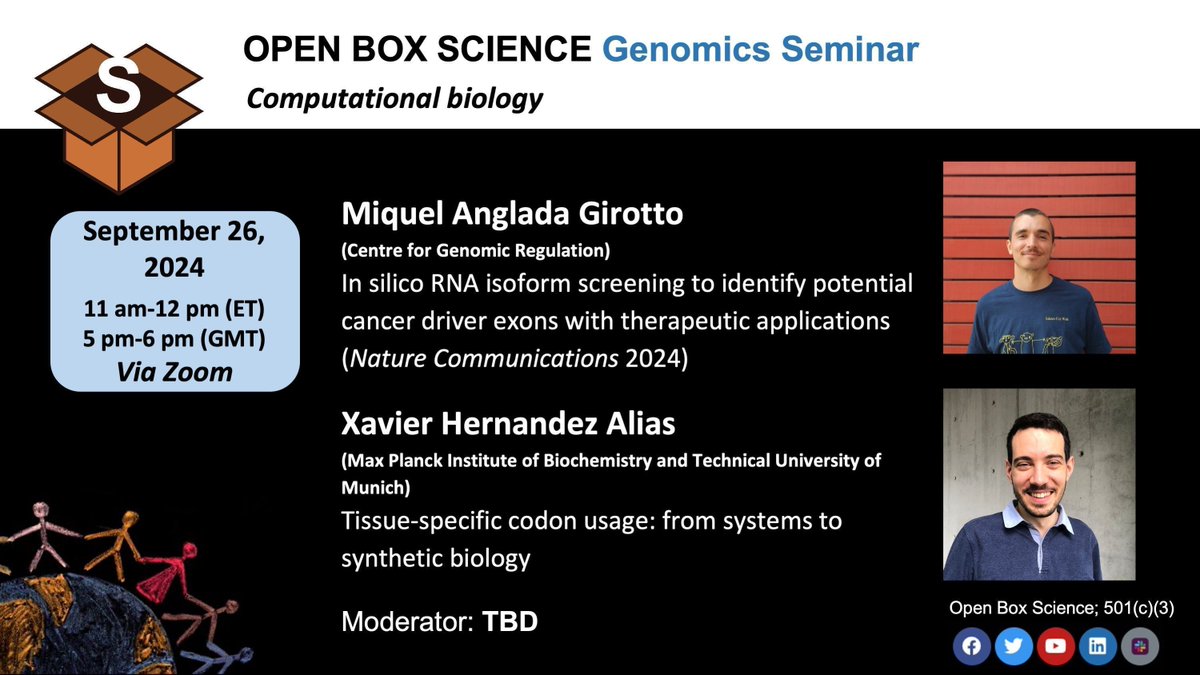
OBS #Genomics Seminar 📆 Thursday, 9/26, 11 am ET/5 pm CET 🎙️ Miquel Anglada Girotto Miquel Anglada-Girotto (m1quelag@bluesky) Centre for Genomic Regulation (CRG) serranoLab 🎙️ Xavier Hernandez Alias Xavier Hernandez Alias Max Planck Institute of Biochemistry (MPIB) gagneurlab 👉 Registration: us02web.zoom.us/meeting/regist… #openscience Moderator: Anna Salamero Boix

DeepRVAT now out! High potential for rare disease res.: a) applies to very imbalanced case-control settings, b) our gene impairment score is trained w/o the pervasive disease-gene list bias: the strength of training on UKBB. Gr8t work Brian Clarke Eva Holtkamp w Oliver Stegle



Information leakage due to homology: A pervasive and non-trivial issue in bioinformatics. Excited to hear about Abdul Muntakim Rafi's insights and recommendations for sequence-based modeling on Wednesday at the Kipoi seminar.

Tomorrow at #ASHG2024, Shubhankar Shubhankar Londhe will present FuncRVP, our model improving rare variant testing by integrating gene impairment scores from DeepRVAT with functional gene information. Don’t miss it: we got fresh results since our July preprint! doi.org/10.1101/2024.0…

