Felix Grünberger
@felixgrunberger
Postdoctoral Researcher at University of Regensburg | Microbiology | Transcriptomics in prokaryotes
ID: 713459142653059072
25-03-2016 20:14:49
54 Tweet
151 Followers
301 Following





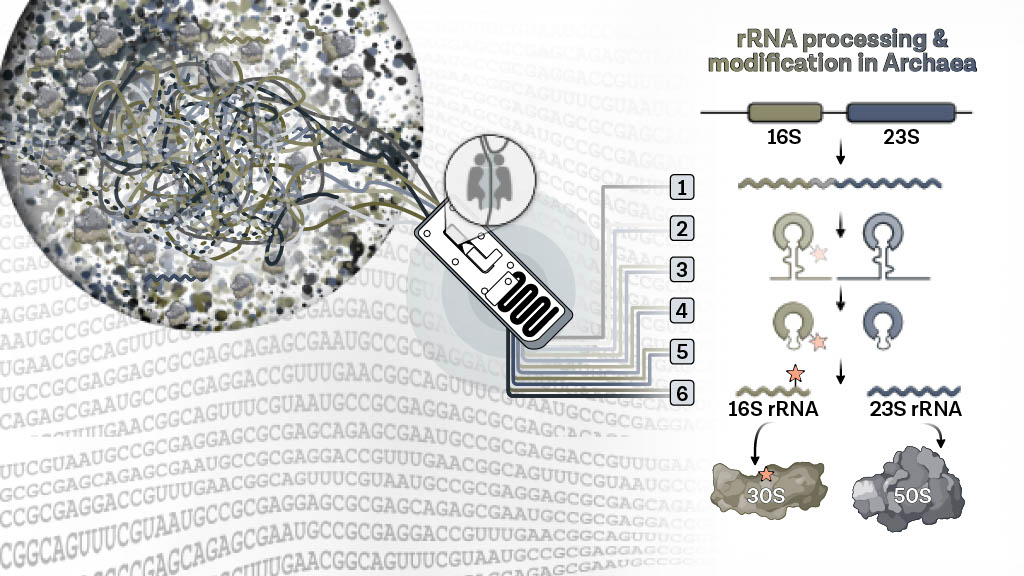
#ResultatScientifique🔎| Explorer les secrets du métabolisme de l'ARN des micro-organismes extrêmophiles 🤝 CNRS 🌍 CNRS à Paris-Saclay SFC_Lab École polytechnique ✍️ Sébastien Ferreira-Cerca et Dina Grohmann 📕 RNA | buff.ly/3O7lJoT buff.ly/3NNBuzX


Working with my physicist colleagues at the Universität Regensburg is a privilege and a lot of fun. The Franz J. Giessibl pushed their high-end qPlus sensor AFM technique to image archaeal S-layers ... here is the result 😍arxiv.org/abs/2307.07052 (just accepted in JPCB). Daniela Tarau


🔥Heat shock response in #Sulfolobus🔥 👉In this new #mbio paper we combine omics-experiments to shed light on underlying regulatory mechanisms! It was a great collaboration between MICR Research Group - VUB, SFC_Lab, Dina Grohmann, Felix Grünberger,...! journals.asm.org/doi/full/10.11… @fwo VUB