Ethan Stancliffe
@ethanstancliff2
ID: 1361364818247245827
15-02-2021 17:20:44
7 Tweet
39 Followers
110 Following

Pleased to share DecoID, our software for deconvolution of metabolomics MS2 data. DecoID mixes ref MS2 in DBs to match experimental data and facilitate metabolite ID. Congrats to developer Ethan Stancliffe & team Miriam Sindelar Michaela Schwaiger-Haber #metabolomics nature.com/articles/s4159…


Great contribution from the Gary Patti lab (Gary was an ugrad in my research lab a while ago)!!! A Workflow to Perform Targeted Metabolomics at the Untargeted Scale on a Triple Quadrupole Mass Spectrometer pubs.acs.org/doi/10.1021/ac…

Happy to share this study on the Warburg effect in cancer. Congratulations to Yahui Wang on her great work! cell.com/molecular-cell…

NEW COMMENT ONLINE: Ethan Stancliffe Ethan Stancliffe & Gary J. Patti Gary Patti share quick tips for re-using #metabolomics data rdcu.be/cYgFY

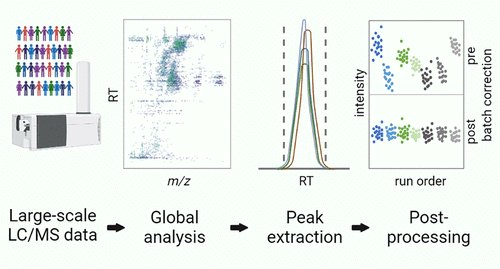
Gary Patti’s lab Gary Patti from WashU Chem developed an untargeted metabolomics workflow designed to support population-based studies using thousands of samples! go.acs.org/3c2

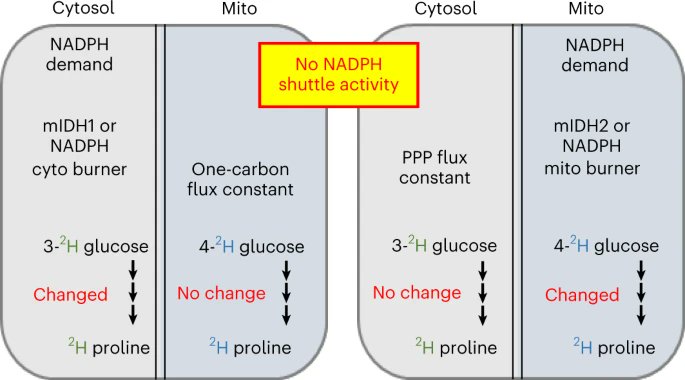
A new paper from Gary Patti and Xiangfeng Niu used glucose tracers and labeling of proline metabolites to assess compartmentalized NADPH fluxes showing that NADPH fluxes in the cytosol and mitochondria are independently regulated. Free to read link at rdcu.be/c8v1r
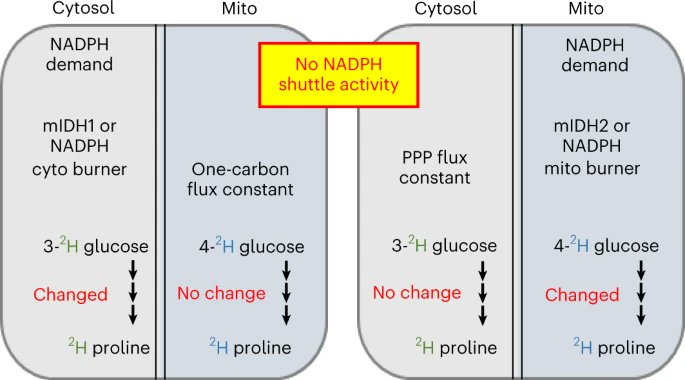

Happy to share new work led by Xiangfeng Niu. We developed a tracing approach to infer changes in cyto & mito NADPH fluxes. Results show that there is no NADPH shuttle in our cells. Wrt NADPH, cyto & mito are on their own. Congrats Niu & team! Nature Chemical Biology rdcu.be/c8v1r