
CompCy Lab @ UNC
@comp_cy
single-cell bioinformatics lab focusing on the immune system! PI: @natstann /in {@unccs,@compmedunc}.
ID: 1358051536937582593
https://compcylab.squarespace.com 06-02-2021 13:55:02
65 Tweet
249 Followers
448 Following

Today is the last day of Alec Plotkin ‘a rotation with us and Justin Milner . He did a great job and is on the way to becoming both an expert of T-cells and graphs. What a great combo of content 🧫👨💻 :) thanks for joining us Alec!






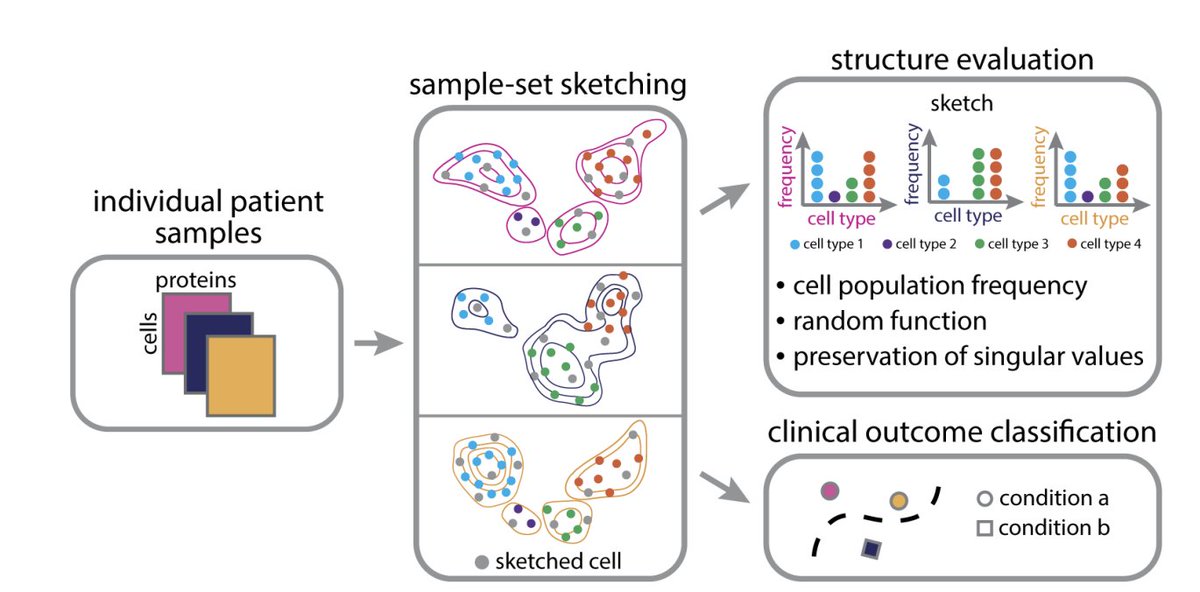
Excited to share our new paper (to appear soon in ACM BCB!) about sketching single-cell data. 😀 Specifically, our sketches preserve cell population frequencies and of course rare populations ;), led by Vishal and Jolene Ranek arxiv.org/abs/2207.00584


.haidyi and Jolene Ranek are in Chicago ACM BCB to present their new papers! 🙂🎉 CytoEMD presented by haidyi : dl.acm.org/doi/10.1145/35… Distribution-preserving sketching presented by Jolene Ranek : dl.acm.org/doi/10.1145/35…

Very proud of Jolene Ranek as her task oriented benchmarking study for integrating RNA velocity and gene expression for trajectory inference and outcome prediction is out today in Genome Biology! genomebiology.biomedcentral.com/articles/10.11… . 🥳

Have you ever wondered if you could leverage RNA velocity data to better resolve biological trajectories or disease phenotypes? Delighted to share that our benchmarking study is out today in Genome Biology! Biggest thank you to Natalie Stanley Purvis Lab 😊 genomebiology.biomedcentral.com/articles/10.11…



Congratulations Alec Plotkin for winning a best poster award at data science day for work predicting CD8 T-cell fates with Justin Milner :) Truthfully, this a picture from last week, but is the same poster. Great job Alec.



Our 🪐SATURN method is now out in Nature Methods! SATURN paves the way for universal cell embeddings, enabling integration of datasets across different species 🐒🐁🧍🐟🐸 Using protein language models, we encode biological meaning of genes in scRNA-seq datasets.







