Christopher Hoel
@chrismhoel
ID: 204113598
17-10-2010 23:30:51
475 Tweet
653 Followers
1,1K Following



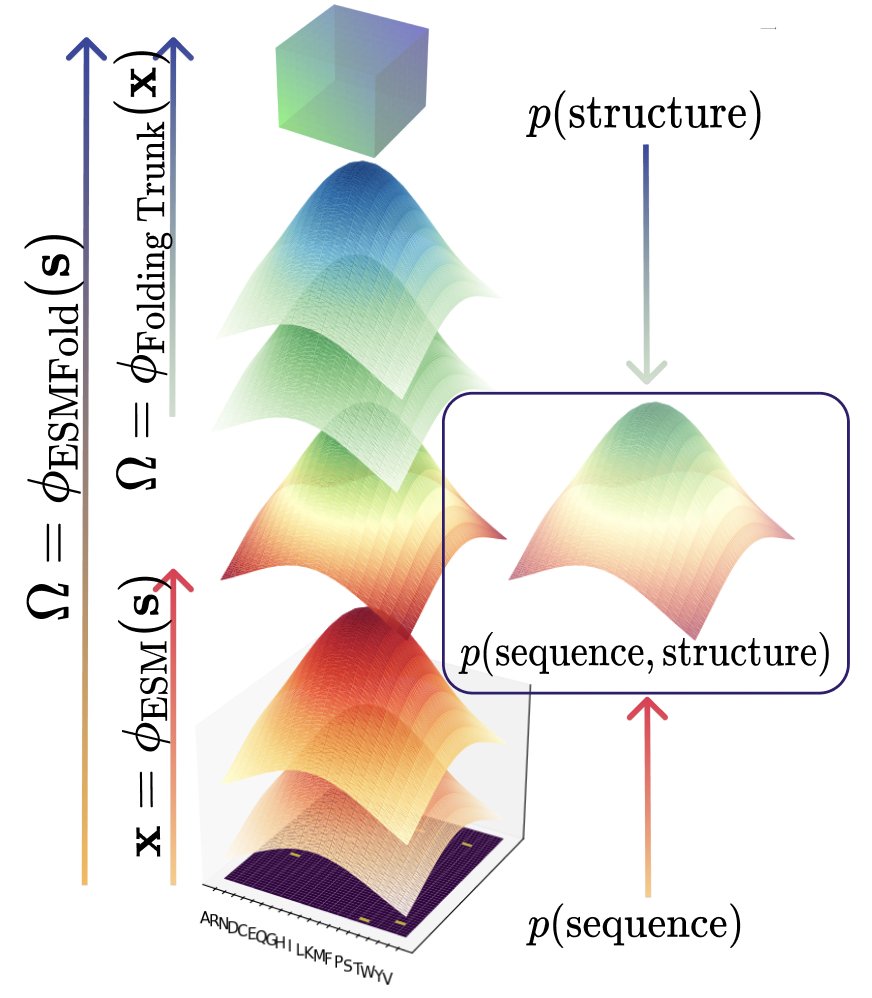
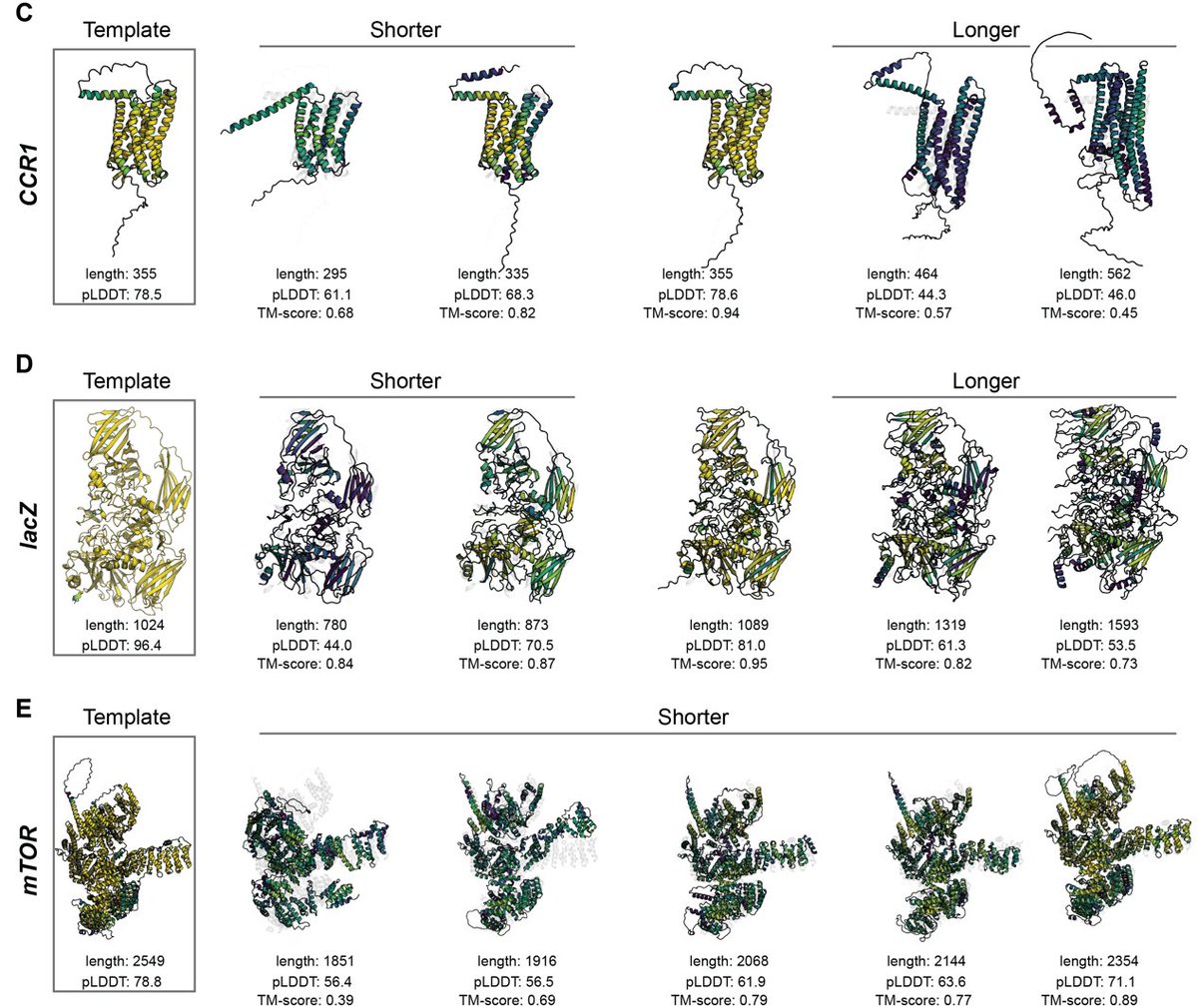
The “known protein universe” is big: hundreds of millions of sequences from thousands of species. Arcadia Science we wondered, how well does it reflect the “actual” protein universe? 🧵[1/15] research.arcadiascience.com/pub/result-pro…


I've noticed that biotech cos built around a computational discovery platform have a different philosophy on company building and how to use software to gain sustainable advantage vs. 'traditional' biotech or pharma. I reflect on this in a new blog post: atelfo.github.io/2024/09/17/a-n…

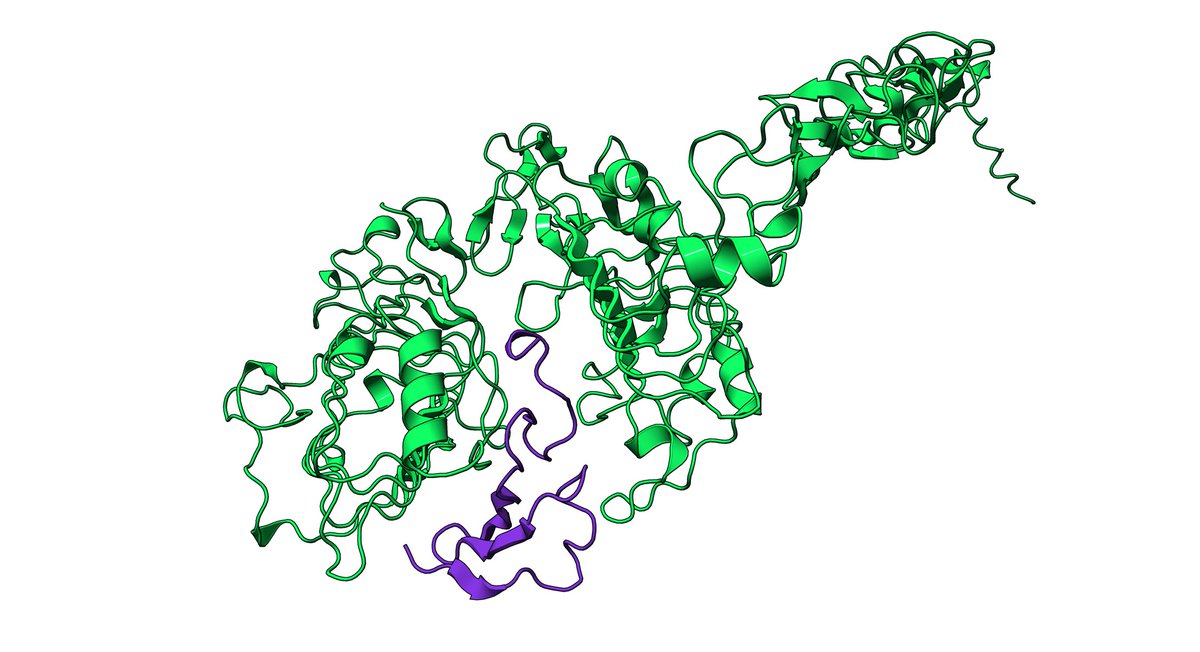
1/ I did my own little hackathon last weekend designing EGFR binders for Adaptyv Bio's protein design competition. I was really excited to see that my submissions took the top 10 spots in the virtual scoring phase! I got some DMs asking about my process so here's a thread:

EvolutionaryScale is partnering with Lux Capital & @envedabio to bring you a Bio x ML Hackathon from Oct 10-20. Build on ESM3 *98 billion* and proprietary datasets of protein activity to develop next-gen bio applications. Open globally. Get started: hackathon.bio



The Adaptyv results are out! Very few designs actually bound. The Design Process text is interesting, e.g., looks like the highest scoring was due to a "new user-friendly binder design pipeline based on AlphaFold2 backpropagation" from Martin Pacesa


Have you ever wanted to design protein binders with ease? Today we present 𝑩𝒊𝒏𝒅𝑪𝒓𝒂𝒇𝒕, a user-friendly and open-source pipeline that allows to anyone to create protein binders de novo with high experimental success rates. Bruno Correia Sergey Ovchinnikov biorxiv.org/content/10.110…

Remember Golden Gate Claude? Etowah Adams and I have been working on applying the same mechanistic interpretability techniques to protein language models. We found lots of features and they’re... pretty weird? 🧵


Can a single small molecule rescue the stability of nearly all mutations in a protein? Our new preprint by Taylor Mighell + thread biorxiv.org/content/10.110…




In new work led by Aditi Merchant with Samuel King, we prompt engineer Evo to perform function-guided protein design with high experimental success rates, including designs that go beyond natural sequences. We also release SynGenome, the first AI-generated genomics database. 🧵 1/N

With the start of my first batch as a visiting partner at Y Combinator, this felt like a good time to write my founder story about our journey at Reverie Labs. It's a bit of a long one. ankitg.me/blog/2025/01/0…