BiomolecularInteractionsLeeds
@bindingleeds
Biomolecular Interactions technology facility at the University of Leeds.
SPR, ITC and MST for binding assays. Refeyn for solution molecular weights.
ID: 1422913653377060866
04-08-2021 13:34:45
96 Tweet
109 Followers
29 Following


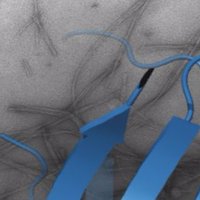
I'm really pleased to have this accepted in Cell Reports A great team effort. Another example of how to use Affimers to understand protein function in cancer. The Astbury Centre University of Leeds FBS Research Facilities Medical Research Council Wellcome cell.com/cell-reports/f…


Biomolecular interactions - our facility technologies. SPR, ITC and MST for affinity, kinetics, thermodynamics and stoichiometry info. Watch this space... #Dianthus NanoTemper



Installation and training on our new Dianthus from NanoTemper Binding assays especially for tricky systems - membrane proteins, multi-molecular complexes and small molecule libraries. Many thanks Medical Research Council for funding. The Astbury Centre @ScienceLeeds #hugthebox






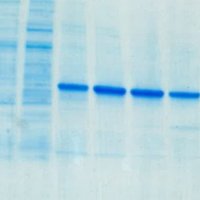
Really pleased the facility could contribute lots of ITC binding results to this publication. Congratulations on your first group publication James F Ross.




Abstract submission for the British Biophysical Society Biennial Meeting 2024 is now open! britishbiophysicss.wixsite.com/swansea2024 Join us for three days of all things Biophysics - and for the first time in Wales! An exciting line-up of speakers, including this year's BBS awardees below:


Bob Schiffrin, Joel Crossley, RadfordLab and colleagues based The Astbury Centre and @ScienceLeeds identify dual binding sites in the outer membrane protein chaperone SurA. #chaperones, #NMR, #smFRET nature.com/articles/s4146…


Excited to have Neo detectors installed on our Fida this week. Extending the concentration range for size, shape, heterogeneity & aggregation measurements. Kinetics & binding assays also from Fidabio. Thanks Tom & Joanne Walter Many thanks Biotechnology and Biological Sciences Research for funding.