
Benno Schwikowski
@bennos
Researcher in #SystemsBiology at @InstitutPasteur
ID: 18160140
16-12-2008 10:45:20
27 Tweet
105 Followers
78 Following

Our lab at Institut Pasteur, since 1887 in Paris has open #postdoc positions at the interface of statistical data analysis and cancer/autoimmune disease. tinyurl.com/ipsysbio #bioinformatics #jobs

Our lab's debut paper in single cell transcriptomics, co-led by our shared PhD student Pierre Bost. Congratulations, Pierre!

Did you ever wonder how well pathway and transcription factor analysis work for scRNA-seq data? Christian Holland (Christian Holland) et al. shed light on this question in our latest preprint [biorxiv.org/content/10.110…]. 1/6
![Saez-Rodriguez Group (@saezlab) on Twitter photo Did you ever wonder how well pathway and transcription factor analysis work for scRNA-seq data? Christian Holland (<a href="/mr_netherlands/">Christian Holland</a>) et al. shed light on this question in our latest preprint [biorxiv.org/content/10.110…]. 1/6 Did you ever wonder how well pathway and transcription factor analysis work for scRNA-seq data? Christian Holland (<a href="/mr_netherlands/">Christian Holland</a>) et al. shed light on this question in our latest preprint [biorxiv.org/content/10.110…]. 1/6](https://pbs.twimg.com/media/EDck5TQXkAIb77B.png)



I am grateful to EU #H2020 for supporting our #ovariancancer research. The main goals of Deciderproject are 1) understand mechanisms causing chemoresistance in #ovariancancer patients and 2) deliver tools for personalized diagnosis and treatment options.

Pleased to inform that our 1st Deciderproject article is published. Fascinating what AI can achieve @precisionpathology. Artificial intelligence-based image analysis can predict outcome in high-grade serous carcinoma via histology alone disq.us/t/41hgk1e

Excellent review on platinum resistance by Huang et al. in Oncogene Journal. Systematic literature search resulted in >900 #PlatinumResistance related genes conveniently stored in a database. #SBLread, Hautaniemi Lab, Deciderproject, Hercules project. rdcu.be/czD19

Register now for this free online training school by TranSYS Institut Pasteur, since 1887 h2020transys.eu/transys-data-a… #dataanalysis #precisionmedicine #itn #school #training



Preclinical models are important tools in #ovariancancer research. We here describe how to generate xenograft models. @medofak_uib Haukeland reachoutinova Gynkreftforeningen @Kreftforeningen Emmet Mc Cormack #CCBIO link.springer.com/protocol/10.10…


Our paper on #scRNAseq analysis of #chemoresistance in #ovariancancer is out today in Science Advances! Many thanks to Sampsa Hautaniemi Anna Vähärautio AnttiHakkinen @e_p_erkan Sanaz Jamalzadeh and all co-authors for your great contribution! science.org/doi/10.1126/sc… (1/3)

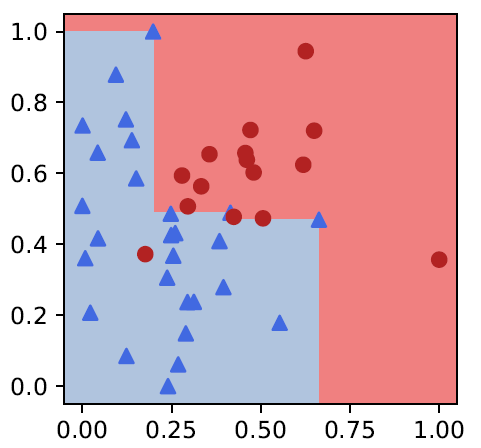
Together with Benno Schwikowski team Institut Pasteur, since 1887 we study ensembles of bivariate monotone classifiers for biomedical applications. In this bioRxiv we present a preprocessing step that speeds up their construction by factor 10-20: doi.org/10.1101/2023.0…

Winter-meeting for the PARIS-project Copenhagen. #ovariancancer Benno Schwikowski, Anna Vähärautio, Wennerberg Lab, Gynkreftforeningen, reachoutinova, PrecOS Lab, Haukeland, Katrin Kleinmanns Norges forskningsråd, @medofak_uib, @eraper (PARIS = Precision drugs Against Resistance In Subpopulations)


Comparative evaluation of feature reduction methods for drug response prediction Scientific Reports 1. This study is the first to compare nine feature reduction (FR) methods for drug response prediction (DRP) on cell line and tumor transcriptomes, using over 6,000 machine learning (ML)



Different levels of #omics data visualized in Hercules project project

