Ashlyn Anderson
@ashlyn_anderso
PhD student @UAB_GBS | Cochran Lab @HudsonAlpha | gene regulation, neurogenomics
ID: 1299419726330892289
28-08-2020 18:53:09
608 Tweet
158 Followers
568 Following











Ever wonder how the genome of human brains compares with macaque, marmoset, mouse, rat, cat, rabbit, horse, and cow? Wonder no longer! Check out a really cool pre-print from my very talented graduate student Ashlyn Anderson on multiomics across mammals: biorxiv.org/content/10.110…

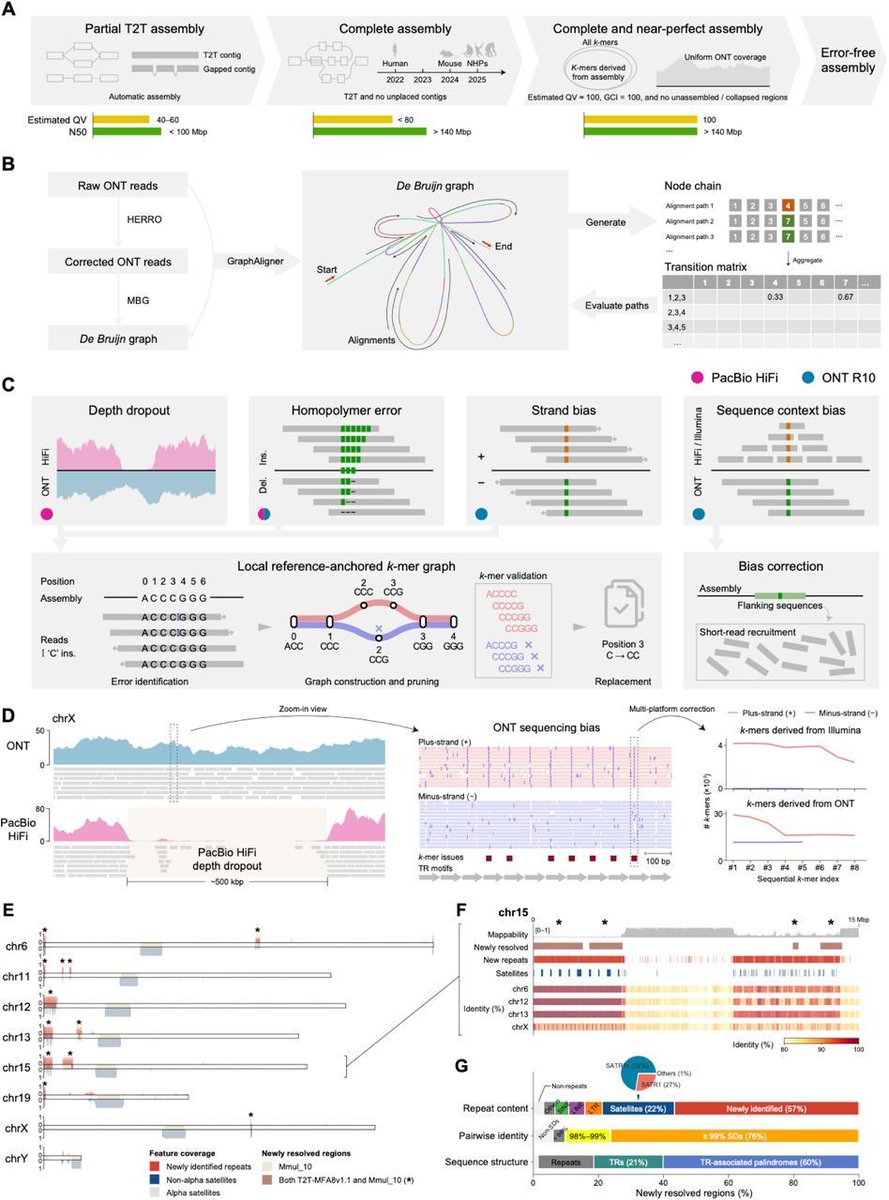
A complete and near-perfect rhesus macaque reference genome: lessons from subtelomeric repeats and sequencing bias. #RhesusMacaque #ReferenceGenome #T2T #Genomics bioRxiv Genomics biorxiv.org/content/10.110…

brilliant work from Johan Jakobsson biorxiv.org/content/10.110…