Androo J. Markham
@androojmarkham
field applications for @nanopore
ID: 1012400448148418560
28-06-2018 18:20:51
154 Tweet
230 Followers
298 Following



Great webinar on Oxford Nanopore ultra long sequencing with great tips @ tricks for human, mammalian and plant species. Thanks all and especially Karen Miga and Adam Phillippy for organising and being great hosts! #T2T


The latest blog post from Science unlocked: publication picks from April 2024 nanoporetech.com/blog/science-u… Oxford Nanopore

We're making several Oxford Nanopore datasets available to coincide with #LC2024 #LondonCalling2024. They are available for download now from the #EPI2ME blog. These include; Zymo fecal reference, plasmids, lambda and E. coli and some human reference RNA! labs.epi2me.io/lc2024-datasets

Impressive recent progress on DNA modification detection from Oxford Nanopore #NanoporeConf

At Oxford Nanopore we have successfully sequenced a yeast chromosome, telomere to telomere, in one read.



We recently updated the Oxford Nanopore #EPI2ME application and a handful of workflows to bring you the IGV browser in app. Bede Constantinides wrote a blog post about this new feature! Check it out: labs.epi2me.io/igv-integration

Curious about sequencing #RNA directly including mod bases? Register to join me for this webinar (11 July) explaining how #directRNA seq works plus recent updates & how they might work for your research needs! I'll be available for Q&A afterwards too. Oxford Nanopore #mRNA Details⬇️




After 4 days the P2S can finally relax after 170 Gbp from one Oxford Nanopore flowcell. No wash! No reload! This is the regular flowcells. Will be looking forward to test the updated chemistry ones at some point.


Introducing NanoCore! A user-friendly software for Nanopore-based genomic surveillance in healthcare facilities. 🖥️Presented in EIC Jack Gilbert, the software's accuracy is equivalent to gold standard short read-based analyses. Read the article: asm.social/28F



Very excited to announce our preprint on adapting the Oxford Nanopore Cas9 workflow for R10 flow cells. This is work from the amazing Veronika Scholz and Veronika Schoenrock and team MGZ - Medizinisch Genetisches Zentrum, and is also my very first senior author paper 🧵biorxiv.org/content/10.110…

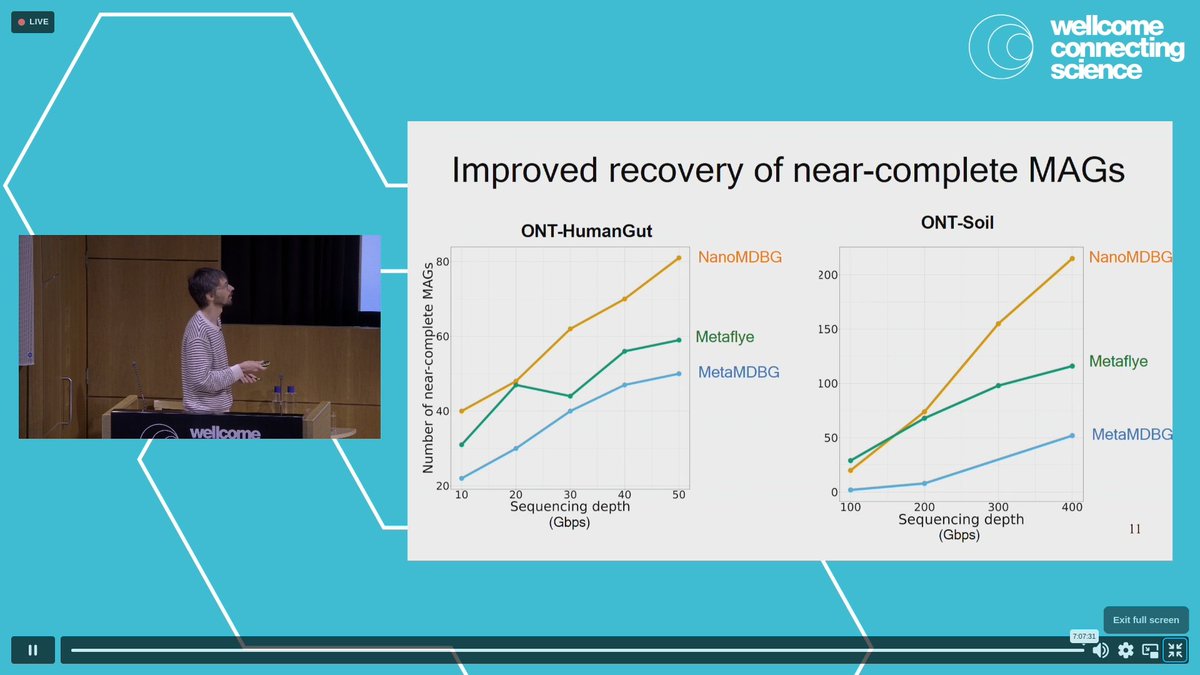
MetaMDBG killing it for #metagenomics assembly! 1. Nicolas Maurice showing how it captures more diversity than metaflye & hifiasm-meta. 2. Gaëtan Benoit Gaetan Benoit presenting new NanoMDBG module that improves it further for Oxford Nanopore #genomeinformatics24 #microbialgenomics


And native barcoding for Oxford Nanopore direct RNA sequencing has arrived ahead of Oxford Nanopore Events. Very impressive first runs Genome Technology Lab with >25 million reads (pre-basecalling) per flow cell. Analyzed data forthcoming!