Alexis Molina
@alexismolinamr
• On the verge of AI 💻 and biology 💊 •
ID: 1494067007905574916
16-02-2022 21:52:11
15 Tweet
48 Followers
570 Following

We are happy to Announce our Latest #SilverSponsor Nostrum Biodiscovery for our Digi-Tech Pharma & AI 2023 Conference to be held on the 10th & 11th of #may this year at #London, UK! #DTP #Digitechpharma Check out the Latest Agenda & Speakers: corvusglobalevents.com/conference/dig…


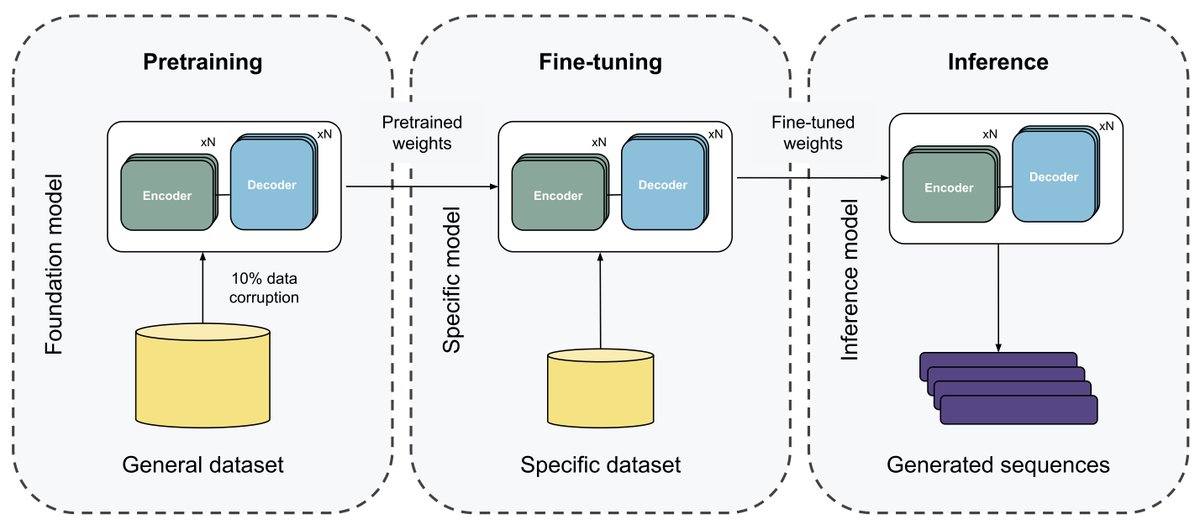
Our latest work at Nostrum Biodiscovery is out! We trained a protein language model of only 14M parameters, a huge downsize in comparison with the latest pLMs. Gen. seqs. have similar properties to the natural ones, show great folding and are stable in MD. Stay tuned for more!






If you want to work on AI projects where pretraining isn’t dead, join a techBio drug discovery company 😜 CC: Ilya Sutskever