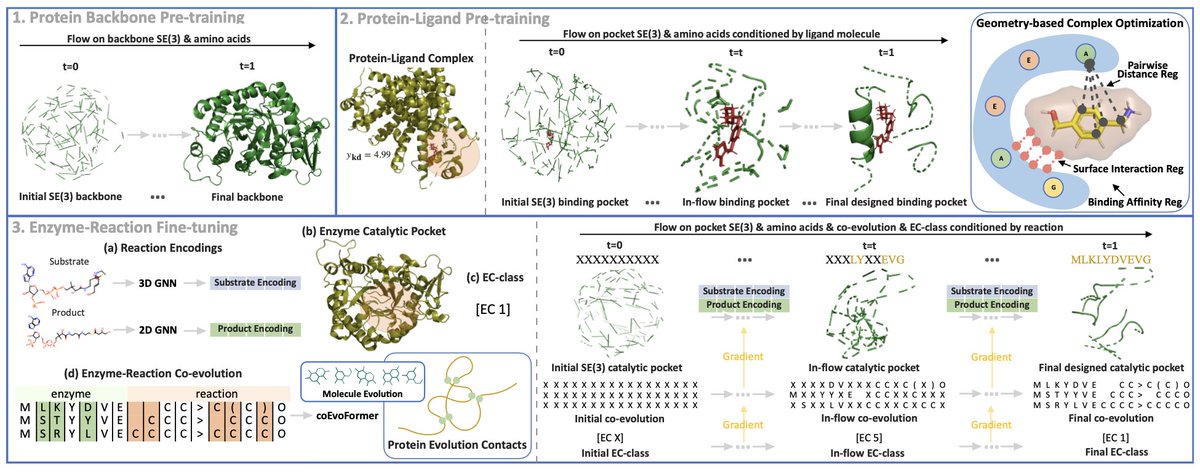
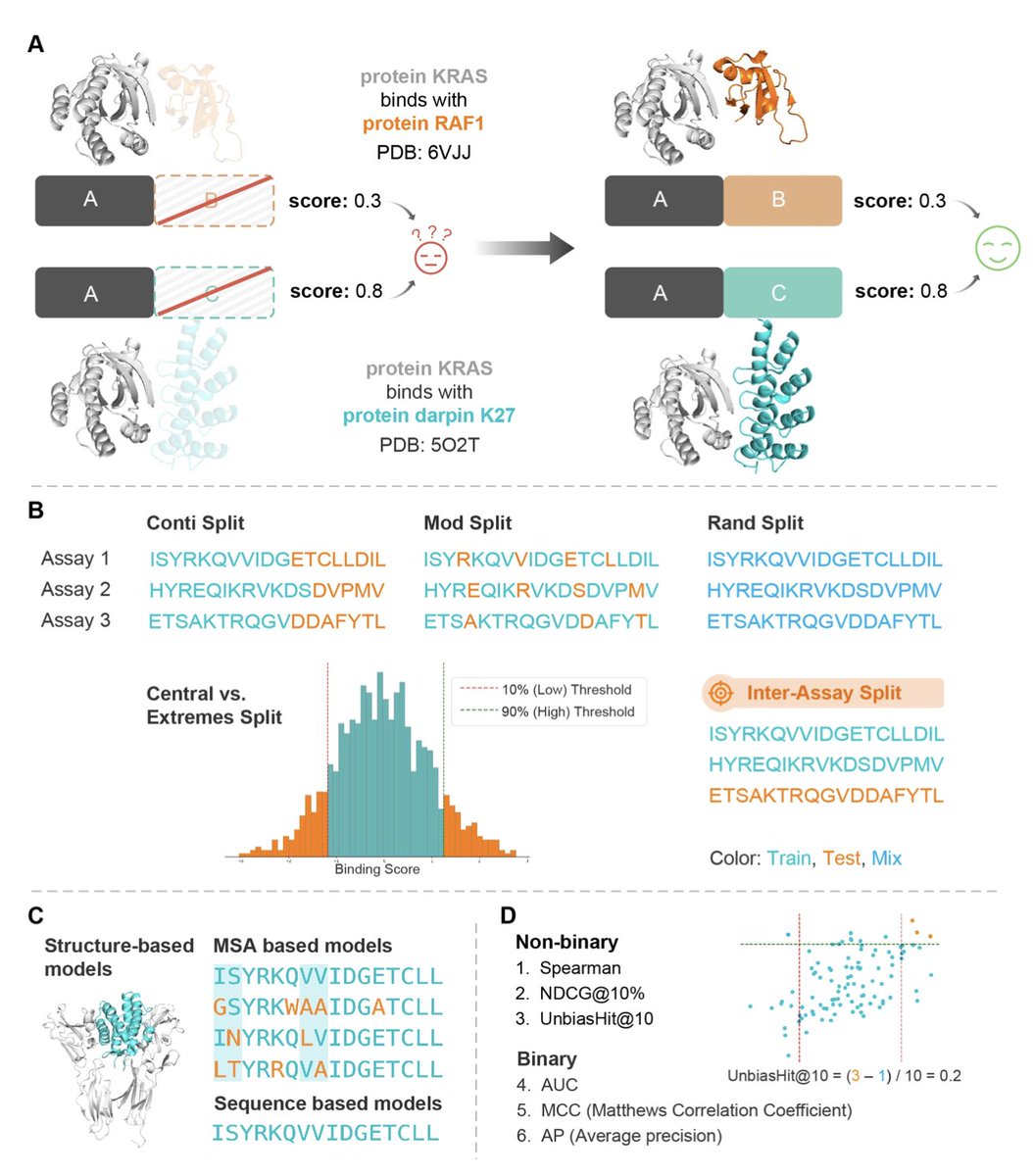
Shuangjia Zheng
@edgar_zheng
Assistant Professor at Shanghai Jiao Tong University (SJTU), Generative AI for Life Science,#DeepLearning #GenerativeAI #DrugDicovery
ID: 822999842334982144
https://zhenglab.sjtu.edu.cn/ 22-01-2017 02:50:27
20 Tweet
72 Followers
84 Following