Dominik Klein
@dominik1klein
@EllisforEurope PhD student @HelmholtzMunich, Student Researcher @Apple. Interested in ML, Single-Cell Genomics, and People.
ID: 1366484427136774151
01-03-2021 20:24:25
61 Tweet
341 Followers
163 Following



GIF art by Théo Uscidda ! Let’s meet and talk :)




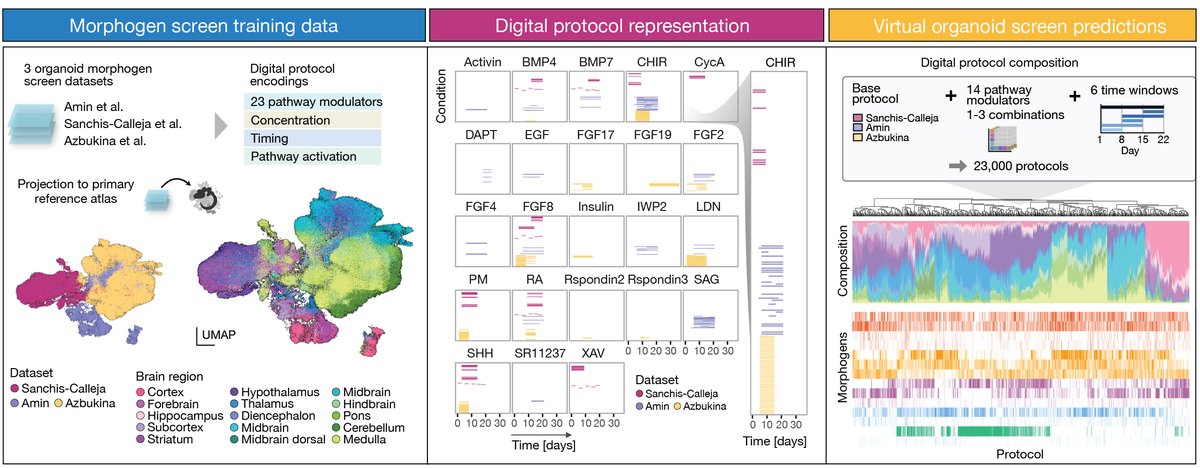
Learned a lot working with Jonas (josch1@bsky) Fabian Theis @dominik1klein Daniil Bobrovskiy Treutlein lab on CellFlow: generative single-cell phenotype modeling with flow matching. Virtual protocol screening is particularly innovative! #organoids Institute of Human Biology Helmholtz Munich | @HelmholtzMunich ETH Zürich