Stuart MacGowan
@stuartmac44
Computational biologist | Protein structures & human variation | R & Python | Genetic effects on protein function | See: go.nature.com/442B3tx
ID: 800932430
https://www.dundee.ac.uk/people/stuart-macgowan 03-09-2012 18:20:06
221 Tweet
442 Followers
1,1K Following

We have released the successor to ANARCI - ANARCII - a suite of Seq2Seq language models trained to number antibody (or TCR) sequences! Read the paper: biorxiv.org/content/10.110… Play with the webtool: opig.stats.ox.ac.uk/webapps/sabdab… Documentation and codebase: github.com/oxpig/ANARCII







MolSnapper has now been published in JCIM & JCTC Journals! MolSnapper integrates expert knowledge into diffusion models for structure-based drug design using a conditioning approach. Congratulations Yael Ziv, Fergus Imrie, Brian Marsden, and Charlotte Deane. pubs.acs.org/doi/10.1021/ac…

We're hiring a Research Software Engineer to join OPIG and Oxford Statistics! This is a permanent role to support the group's world-leading open source tools Grade 8 salary band £48k-£57k pa. Apply by 4th June 2025, noon UK time. Further details here: my.corehr.com/pls/uoxrecruit…



New publication. We quantify the aggregation of >100,000 random protein sequences to train CANYA, a convolution-attention hybrid neural network to predict aggregation from sequence. With Benedetta Bolognesi Centre for Genomic Regulation (CRG) Wellcome Sanger Institute IBEC science.org/doi/10.1126/sc…

Come and join us! We’re hiring a new Group Leader in Generative Biology at the Wellcome Sanger Institute Building AI models or the data to train them? Core funding of >$130M a year for a faculty of ~30. pls RT! nature.com/naturecareers/… acrobat.adobe.com/id/urn:aaid:sc…


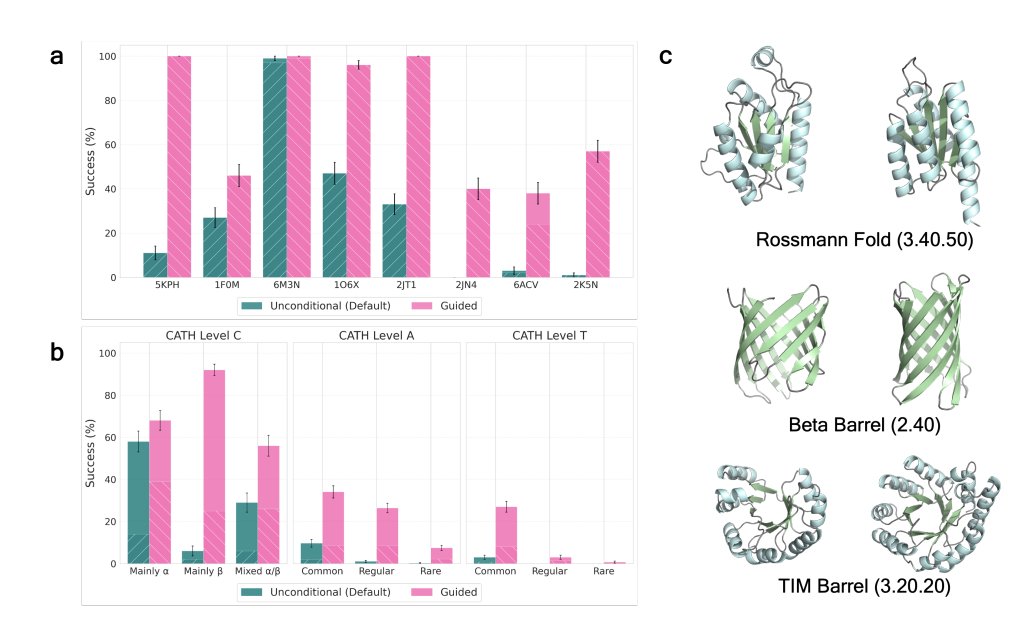
ProteinGuide: Predictor guidance for generation with any autoregressive or discrete diffusion protein model. Junhao (Bear) Xiong Jennifer Listgarten