
Chayaporn Nok Suphavilai
@nokcs
ID: 45359327
07-06-2009 15:39:11
19 Tweet
61 Followers
121 Following

Our new framework, CaDRReS-Sc, for predicting cancer drug response in heterogeneous tumors based on single-cell data. github.com/CSB5/CaDRReS-Sc biorxiv.org/content/10.110… I’m so grateful for the support and guidance from Niranjan Nagarajan Rishab Dasgupt Ankur Sharma :)

We have some "strong" transfer learning mojo for you ... w/ Rafael Peres Chayaporn Nok Suphavilai TUGDA: task uncertainty guided domain adaptation for robust generalization of cancer drug response prediction from in vitro to in vivo settings academic.oup.com/bioinformatics…

What a pleasant surprise to see this out in Genome Medicine! Great collaboration with Chayaporn Nok Suphavilai Ankur Sharma Ramanuj Dasgupta & Shumei Chia




Chayaporn Nok Suphavilai, Karrie Ko : Sentinel-site sequencing in near real-time for detecting clonal outbreak clusters and providing alerts. Our new case study with Oxford Nanopore sequencing for whole-genome characterization of Shigella flexneri isolates frontiersin.org/articles/10.33…

We integrated raw inconsistent drug response data to build an integrative pharmacogenomics database. CREAMMIST provides easy-to-use statistics and uncertainty info for various downstream analyses, such as identifying biomarkers and machine learning models. academic.oup.com/nar/advance-ar…

Discovery of the sixth Candida auris clade in Singapore medrxiv.org/content/10.110… We are excited to share our recent discovery of the sixth major Candida auris clade in Singapore Diag Bacteriology Chayaporn Nok Suphavilai Jacquomo Niranjan Nagarajan Clement Tsui 徐健銘

Brilliant work and collaboration with Chayaporn Nok Suphavilai Karrie Ko Jacquomo Niranjan Nagarajan Discovery of the sixth Candida auris clade in Singapore medrxiv.org/content/10.110…


The MetageNN paper is finally out on bioRxiv! MetageNN outperforms recent machine learning based approaches for taxonomic classification, and shows higher sensitivity than kmer based tools for novel taxa ... w/ Rafael Peres Chayaporn Nok Suphavilai biorxiv.org/content/10.110…

MetageNN - a memory efficient taxonomic classifier - is now out in BMC Bioinformatics #metagenomics #AI MetageNN outperforms other machine learning-based metagenomic classifiers, and shows higher sensitivity than kmer-based tools Rafael Peres Chayaporn Nok Suphavilai tinyurl.com/mvr5th8j

Check out our paper now on The Lancet Microbe ! We Chayaporn Nok Suphavilai Niranjan Nagarajan Jacquomo Clement Tsui 徐健銘 made the discovery in June 2023 and made the data available for the community as a preprint in August 2023. Now it is finally here! thelancet.com/journals/lanmi… #candidia #candidaauris #Genomics

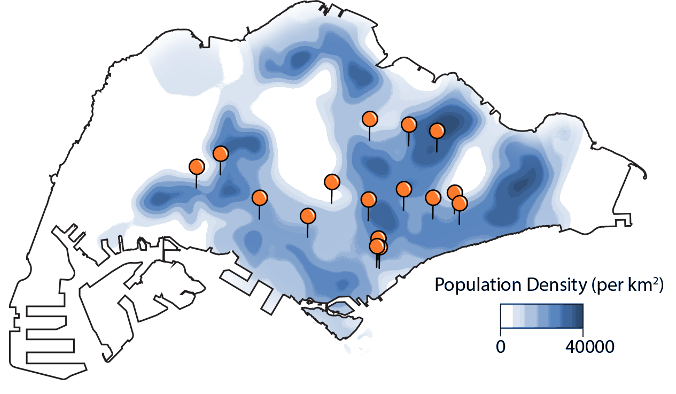
Excited to share our work on city-wide metagenomic surveillance of hawker centres in Singapore! w/Macadology Several surprising findings here ... see thread medrxiv.org/content/10.110…

Senior scientist at A*STAR Genome Institute of Singapore (A*STAR GIS) Dr. Chayaporn Suphavilai Chayaporn Nok Suphavilai is one of Singapore's Top 100 Women in Tech 2023 honorees. She is passionate about applying genomic sequencing, analyses, and artificial intelligence techniques to make tangible differences in the healthcare domain.



Find out how our researchers at A*STAR Genome Institute of Singapore (A*STAR GIS)—Associate Director Niranjan Nagarajan, Scientist and GIS Innovation Fellow Chayaporn Nok Suphavilai and Dr Karrie Ko—discovered a new variation of 𝘊𝘢𝘯𝘥𝘪𝘥𝘢 𝘢𝘶𝘳𝘪𝘴 (𝘊.𝘢𝘶𝘳𝘪𝘴)—𝗰𝗹𝗮𝗱𝗲 𝗩𝗜—a drug-resistant yeast that kept infectious disease
