Arang Rhie
@arangrhie
Staff Scientist in Genome Informatics
ID: 817382322274070528
06-01-2017 14:48:26
482 Tweet
1,1K Followers
146 Following



I am thrilled to share our study on the de novo assembly and characterisation of 43 diverse human Y chromosomes, published today in nature, together with the first #T2T Y assembly from Adam Phillippy, Arang Rhie and co-authors nature.com/articles/s4158…


Our preprint is out! We hacked the Oxford Nanopore sequencer to read amino acids and PTMs along protein strands. This opens up the possibility for barcode sequencing at the protein level for highly multiplexed assays, PTM monitoring, and protein identification! biorxiv.org/content/10.110…





Genome Research is now encouraging submissions for a Special Issue on Long-read DNA Sequencing Applications in Biology and Medicine, Guest Edited by AnaConesa, Alexander Hoischen, and Fritz Sedlazeck bit.ly/grcallforpapers #longreads #T2T #LongTREC #Hitseq #ASHG23


Just in time following an incredible #ASHG23, we're excited to share our work on quantifying sources of gene expression variation across a global human cohort! At the helm of this project is Dylan Taylor in Rajiv McCoy's lab! biorxiv.org/content/10.110…

So happy to finally see our MAGE paper out on bioRxiv!! I'm so excited about this data set; I hope this will be a valuable resource for the field of functional genomics. Congrats as well to my co-authors Surya Chhetri, Michael Tassia, @aabiddanda, Alexis Battle, and Rajiv McCoy

I am excited to share that National Human Genome Research Institute has created a new publicly available digital archive and search aid for accessing documents related to the history of genomics!



Preprint on Exploring gene content with pangenome gene graphs: arxiv.org/abs/2402.16185. It describes pangene for building gene graphs and for calling gene-level variations which can be found at pangene.bioinweb.org. Pleasant collaboration with Maximillian Marin and Maha Farhat.


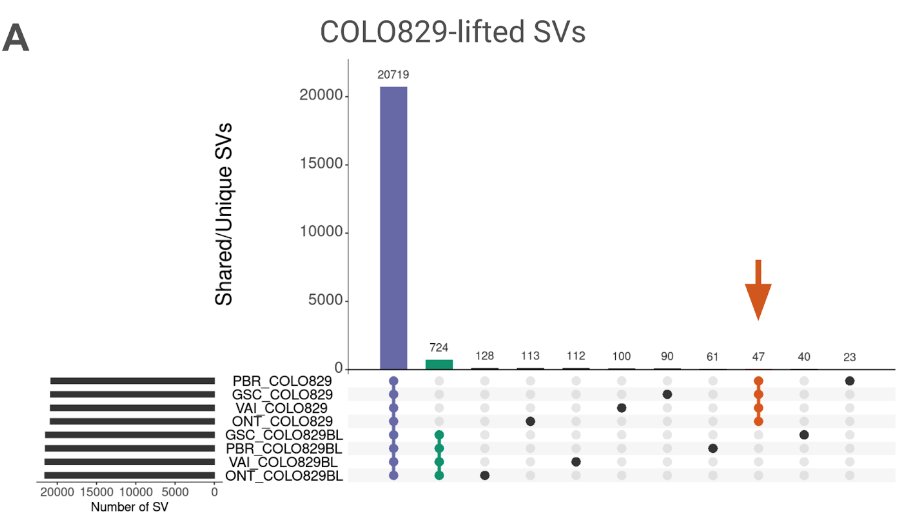
New work on #Cancer variant prioritization using #T2T . It helps & we can overcome variant annotation issues! + An updated somatic SV benchmark for COLO829/COLO829BL across 4 replicates Grch38 +T2T! New preprint: medrxiv.org/content/10.110… Luis Paulin, PhD BCM HGSC Kieran O'Neill etc!