
Jim Horne
@_jimhorne
Postdoc in the Luisi group at Cambridge Biochem working on the structural biology and biochemistry of bacterial efflux pumps.
ID: 3132864175
02-04-2015 19:36:33
119 Tweet
176 Followers
305 Following

Check out our Cell Preview summary of Dr Matthew Doyle Harris Bernstein 's fantastic paper out now in Cell and learn about why these are super cool structures you should pay attention to! cell.com/cell/fulltext/…

Great tree-of-life-wide OMP bioinformatics paper and tools from Joanna Slusky 🫖. The kind of work I've always thought would be really cool to do on today's huge genome databases! biorxiv.org/content/10.110…



Farewell to single-well #smFRET. Our newest story on bioRxiv biorxiv.org/content/10.110… Great team work led by Andreas with Koushik Sreenivasa, Mathias, @nehagc18, Philipp and Georg Krainer. Brief tweetorial and open access software 1/5


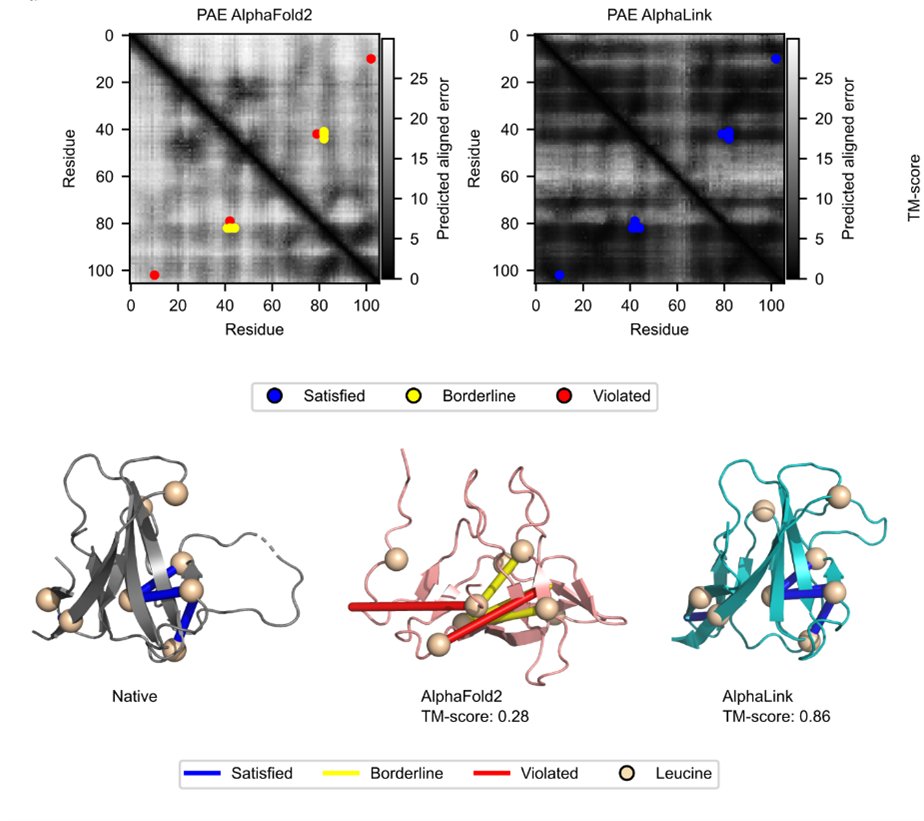
We and the RBO Lab - TU Berlin are excited to present AlphaLink nature.com/articles/s4158… a modified version of the AlphaFold2 protein structure prediction algorithm that can directly incorporate experimental residue-residue contact data from crosslinking MS (and potentially other techniques)!






In our just accepted paper in Angewandte Chemie with RadfordLab & Neil Ranson in The Astbury Centre we used In-Cell PELDOR/DEER complemented by CryoEM to show the BAM complex adopts a lateral closed conformation in E coli cells in presence of antibiotic darobactin-B: onlinelibrary.wiley.com/doi/10.1002/an…

Our new paper published in Journal of Biological Chemistry. Categorization of Escherichia coli Outer Membrane Proteins by Dependence on Accessory Proteins of the β-barrel Assembly Machinery Complex jbc.org/article/S0021-…

We're excited to share new tools, using photo-crosslinking mass spec 💡⚔️⚖️ to study protein structures and complexes in situ with high spatial and temporal resolution. Out now in American Chemical Society ACS Analytical Chemistry Journals. Short 🧵 1/8 pubs.acs.org/doi/10.1021/ac…




New paper from Sujeet Kumar and co, finally finding the OM clamp for one of the non-TamB AsmA-like proteins! If we could just do all vs. all cell envelope proteins in AF2 I wonder what else we would discover pnas.org/doi/full/10.10…


