
Daniel Figeys
@danielfigeys
University of Ottawa.
Microbiome, metaproteomics, metabolomics, metagenomics, bioinformatics, drug- microbiome interactions, precision microbiome nutrition
ID: 745528422764445701
22-06-2016 08:06:42
1,1K Tweet
1,1K Followers
1,1K Following

Huge thanks to all the collaborators at University of Lille (Dr. Trottein team), University of Ottawa (Dr. Figeys team Daniel Figeys), Health Canada (Dr. Sean Li team), and University of Campinas (Dr. Vinolo team).

Excited to be part of the proteomic navigator of the human body (π-HuB). This will transform our understanding of biological processes in human, including the human gut #microbiome . nature.com/articles/s4158… NorthOmics lab

Interested in studying the functions of the microbiome?Check out PhyloFunc, a novel functional beta-diversity metric that incorporates #microbiome phylogeny to inform on #metaproteomic functional distance. NorthOmics lab microbiomejournal.biomedcentral.com/articles/10.11…



Metaproteomic network. Sun et al. analyzed peptide correlations from a large-scale #metaproteomics study. Results: better coverage at genome levels and new links between known and uncharacterized proteins. NorthOmics lab biorxiv.org/content/10.110…


Today marks my last day at Université d'Ottawa | University of Ottawa. Grateful for the years of collaboration, and growth. From founding the OISB to launching the School of Pharmaceutical Sciences—what a journey. Thank you to all who were part of it and to my amazing lab. On to the next chapter! 🚀 NorthOmics lab

An article published in Microbiome presents PhyloFunc: a novel functional beta-diversity metric that incorporates microbiome phylogeny to inform on metaproteomic functional distance. microbiomejournal.biomedcentral.com/articles/10.11…


Interested in #drugs, #microbiome and #antibiotic resistance, check out our paper on the effects of >300 drugs on the gut microbiome. As well, exciting results on antibiotic resistance defining how microbiomes respond to drugs. NorthOmics lab rdcu.be/eMmcT

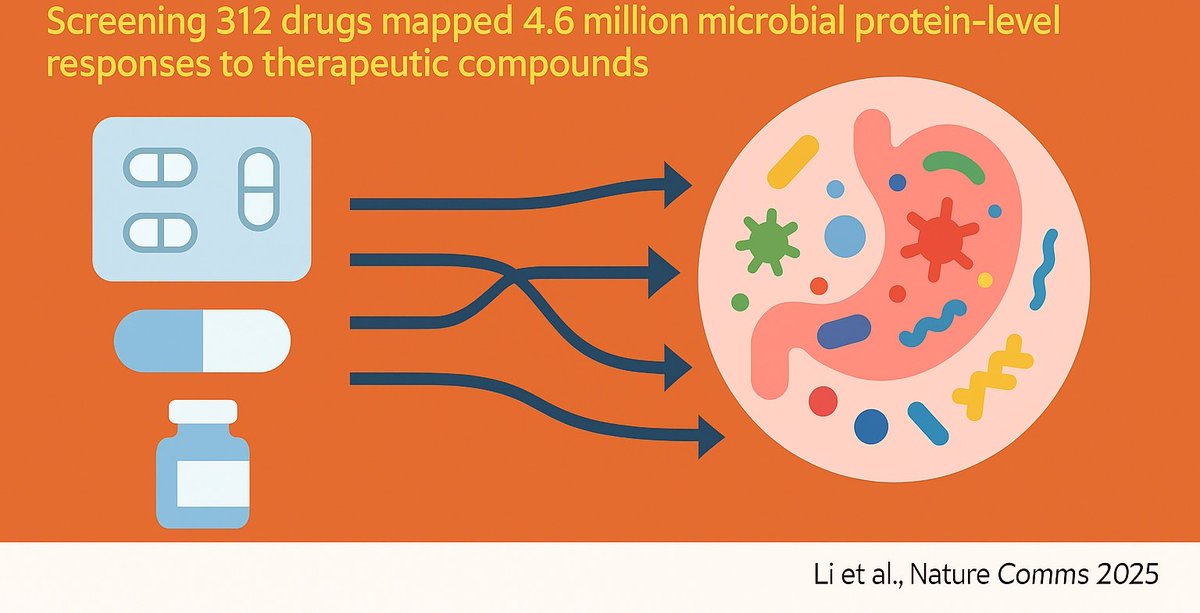
From #drug cabinet → gut #microbiome: screening 312 therapeutic compounds, we mapped 4.6 million microbial protein-level responses to reveal how our drugs don’t just target us, they hit our gut microbes too. NorthOmics lab Quadram Institute rdcu.be/eMmcT

#drugs for the nervous system (#antidepressants/#antipsychotics) stand out: #SSRI/ #SNRI/ #TCA push the gut #microbiome into weaker alternate states, lowering functional redundancy and ramping up #antibiotic-resistance proteins. NorthOmics lab Quadram Institute

Functional redundancy matters: the more overlapping microbial functions you have, the more resilient your #microbiome. We saw a strong negative correlation between redundancy loss and increase in #antimicrobial resistance protein upon #drug treatment of microbiomes NorthOmics lab