
Daniel Seaton
@danieldseaton
Research scientist in early stage drug discovery in Computational Biology dept. @gsk. Views expressed are my own.
ID: 309104298
01-06-2011 14:52:39
462 Tweet
409 Followers
669 Following





Sorry to be missing #eshg2024. From my group, Mahmoud Koko Musa will be presenting a poster on sex differences in autism at 3.45pm today, and Daniel Malawsky is talking at 11.30am on Tuesday in the brain phenotypes session about longitudinal genetic effects on cognition. Do go!


Our lab, in collaboration with partners from Genentech, Sanofi, @gsk, Pfizer Inc., Wellcome Sanger Institute, and EMBL-EBI, has been awarded £2M by Open Targets to generate novel perturbation data across multiple disease models and develop large generative models to facilitate the discovery



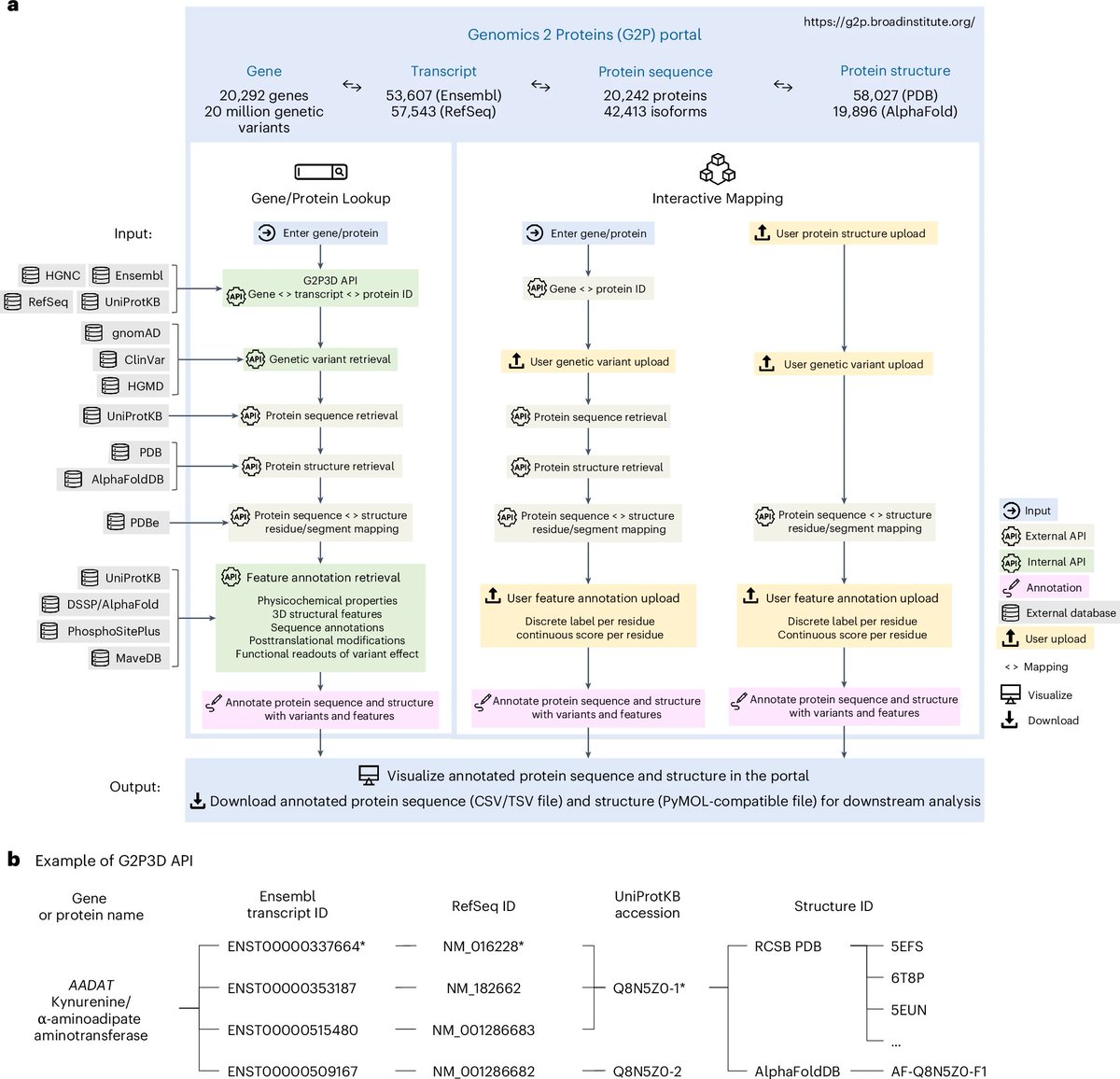
Genomics 2 Proteins portal: a resource and discovery tool for linking genetic screening outputs to protein sequences and structures | Nature Methods - Genomics 2 Proteins (G2P) portal, linking 20,076,998 genetic variants to 42,413 protein sequences and 77,923 structures (58,027



Quantgen friends: we are starting to accept applications for quantgen Gordon Research Conferences Feb 2025, and we’re now 1/3 full! We will make acceptances monthly, and prioritise those with abstracts, so pls submit yours early (you can edit it after submission)! grc.org/quantitative-g…


Probably the most important paper I will ever write link.springer.com/article/10.118… Stephen Burgess BMC Medicine




Are you a postgraduate student interested in protein modelling and drug discovery? We have an exciting opportunity to join our team at GSK for a 6-9 months internship, working on an ambitious cross-department research project. Apply before March 14th! gsk.wd5.myworkdayjobs.com/en-US/GSKCaree…




