Daniel Chang
@danielchang2002
Genetics PhD student @Stanford | Prev @UMNComputerSci | Genomics + ML
ID: 1058949309025132544
http://danielchang2002.github.io 04-11-2018 05:09:24
55 Tweet
178 Followers
262 Following


Excited to announce that I’m starting a new position as a senior fellow FutureHouse. I’ll be working on applying AI agents to metagenomic workflows. Very grateful for the opportunity to learn and explore my scientific interests. Please reach out if you’d like to collaborate!








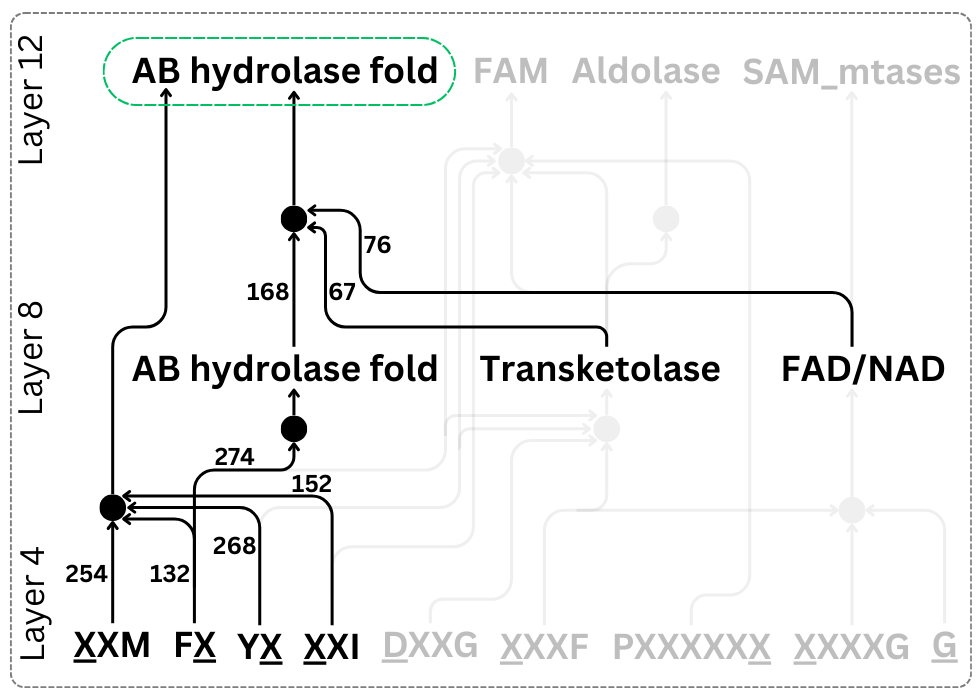
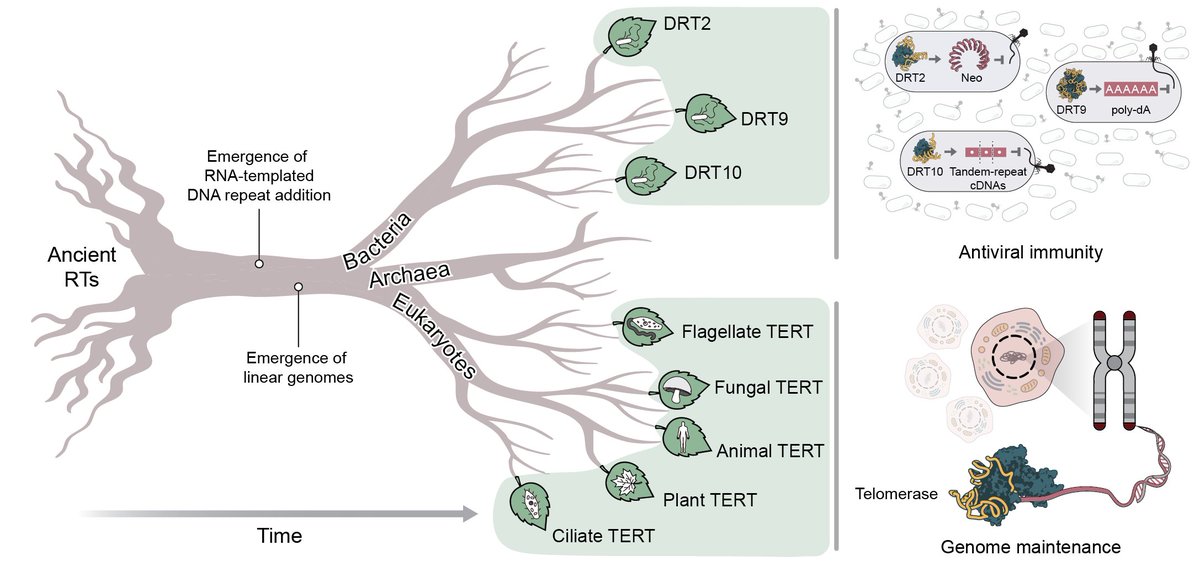
Welcome to the age of generative genome design! In 1977, Sanger et al. sequenced the first genome—of phage ΦX174. Today, led by Samuel King, we report the first AI-generated genomes. Using ΦX174 as a template, we made novel, high-fitness phages with genome language models. 🧵
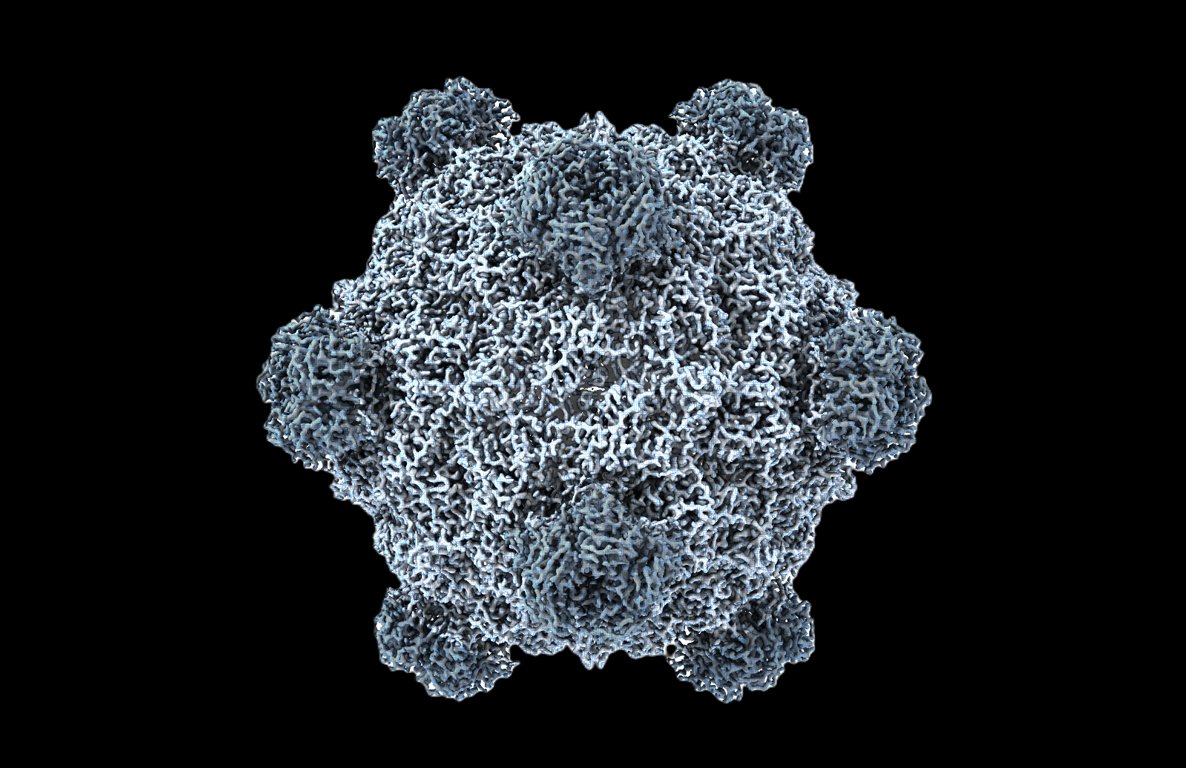

Had fun solving the cryo structure of Evo-Φ36 with the awesome team of Samuel King, Claudia Driscoll, and Max Wilkinson! Samuel is a beast and the reason there is a 'skevophage' react on Stanford slack.



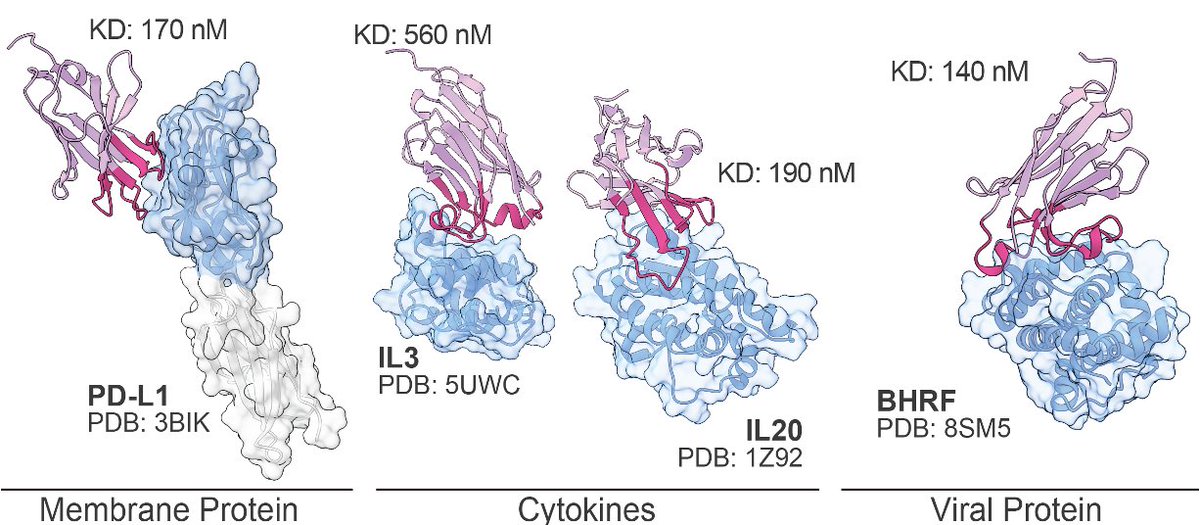
Today, we report Germinal, a method for efficient de novo antibody design, with Santiago Mille and Xiaojing Gao. Germinal achieves success rates of 4-22% across diverse epitopes. We make the work fully open, without doing lame things like posting a preprint without methods. 🧵


Super excited to share what we’ve been working on in collaboration with Xiaojing Gao over the past few months on de novo antibody design. Check out this great thread by our team lead Santiago Mille highlighting the technical aspects of the pipeline!