
NENG HUANG
@csuhuangneng
bioinformatics
ID: 1034689535811055616
29-08-2018 06:29:43
38 Tweet
162 Followers
190 Following





Excited to share our new t2t assembly algorithm for diploid and polyploid genomes! Using 132 assembled haplotypes from the Human Pangenome Reference Consortium , we show that our approach is cost-efficient, robust, and could achieve t2t assemblies without high coverage reads arxiv.org/abs/2306.03399



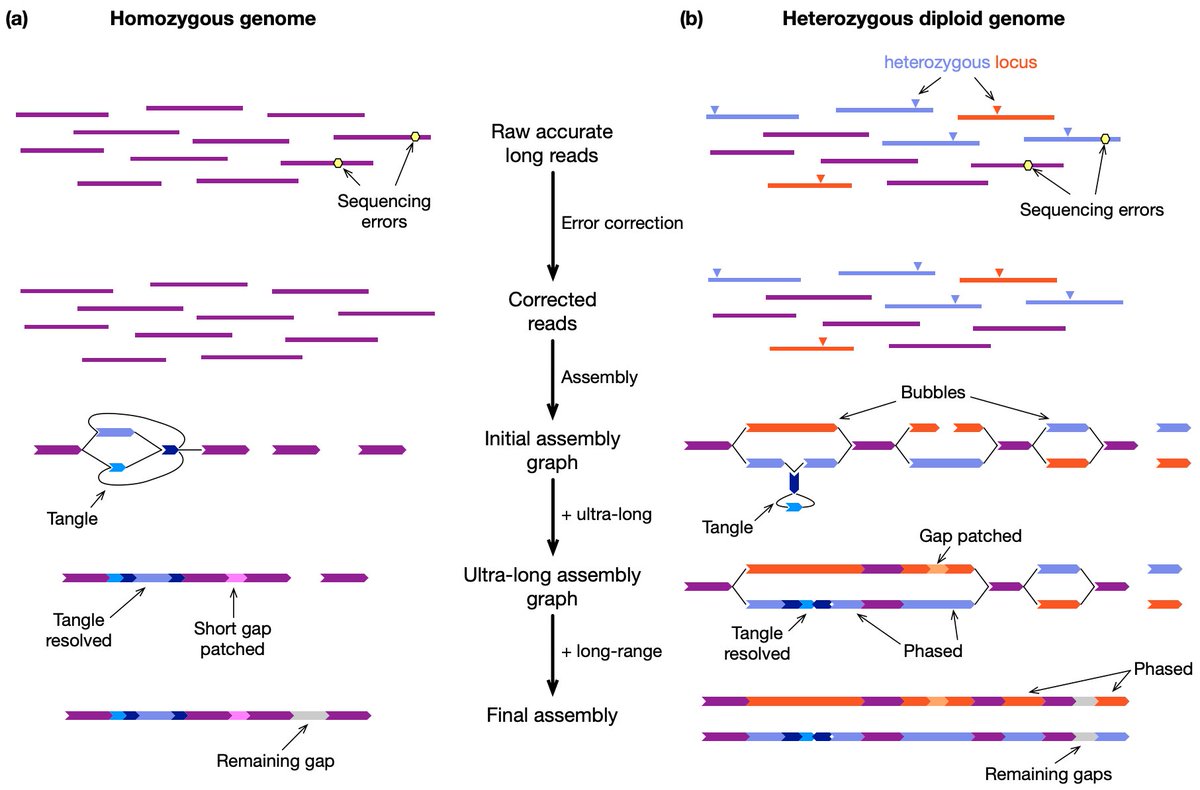
Richard Durbin and I have been working on a review titled "Genome assembly in the telomere-to-telomere era". We have put the preprint at arxiv.org/abs/2308.07877. Let us know if you have comments or suggestions. We would really appreciate!




Excited to share our latest research published in Nature Communications ! Check out our paper on [transposable element detection] here: nature.com/articles/s4146…. Dive into our code and data on GitHub: github.com/CSU-KangHu/HiTE.




Excited to announce Splam is now published in Genome Biology! Splam, trained on thousands of splice junctions from GTEx, is your go-to tool for scoring splice junctions from alignment or annotation files. Mihaela Pertea Steven Salzberg 💙💛 Alan Mao doi.org/10.1186/s13059…




